Glucan Binding Protein C of Streptococcus mutans Mediates both Sucrose-Independent and Sucrose-Dependent Adherence.
Mieher, J.L., Larson, M.R., Schormann, N., Purushotham, S., Wu, R., Rajashankar, K.R., Wu, H., Deivanayagam, C.(2018) Infect Immun 86
- PubMed: 29685986
- DOI: https://doi.org/10.1128/IAI.00146-18
- Primary Citation of Related Structures:
5UQZ, 6CAM - PubMed Abstract:
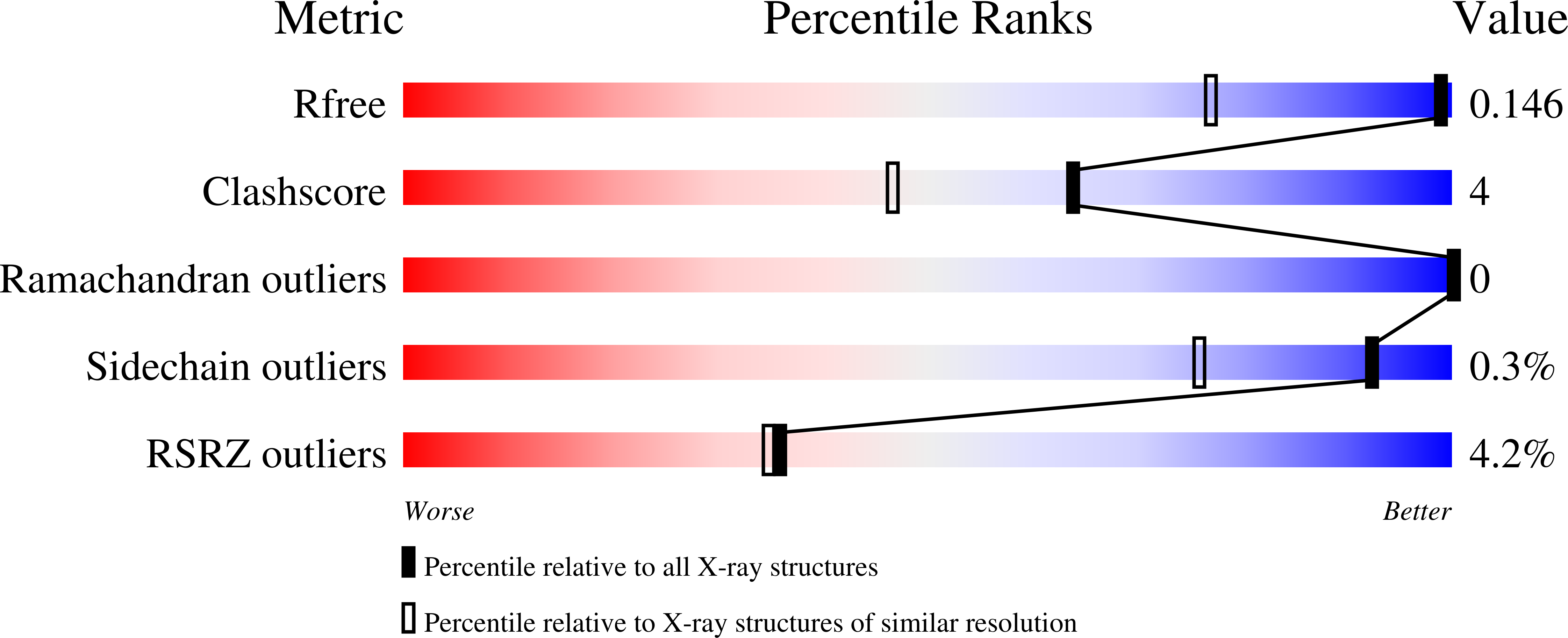
The high-resolution structure of glucan binding protein C (GbpC) at 1.14 Å, a sucrose-dependent virulence factor of the dental caries pathogen Streptococcus mutans , has been determined. GbpC shares not only structural similarities with the V regions of AgI/II and SspB but also functional adherence to salivary agglutinin (SAG) and its scavenger receptor cysteine-rich domains (SRCRs). This is not only a newly identified function for GbpC but also an additional fail-safe binding mechanism for S. mutans Despite the structural similarities with S. mutans antigen I/II (AgI/II) and SspB of Streptococcus gordonii , GbpC remains unique among these surface proteins in its propensity to adhere to dextran/glucans. The complex crystal structure of GbpC with dextrose (β-d-glucose; Protein Data Bank ligand BGC) highlights exclusive structural features that facilitate this interaction with dextran. Targeted deletion mutant studies on GbpC's divergent loop region in the vicinity of a highly conserved calcium binding site confirm its role in biofilm formation. Finally, we present a model for adherence to dextran. The structure of GbpC highlights how artfully microbes have engineered the lectin-like folds to broaden their functional adherence repertoire.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, Birmingham, Alabama, USA.