Critical Role of Water Molecules in Proton Translocation by the Membrane-Bound Transhydrogenase.
Padayatti, P.S., Leung, J.H., Mahinthichaichan, P., Tajkhorshid, E., Ishchenko, A., Cherezov, V., Soltis, S.M., Jackson, J.B., Stout, C.D., Gennis, R.B., Zhang, Q.(2017) Structure 25: 1111-1119.e3
- PubMed: 28648609
- DOI: https://doi.org/10.1016/j.str.2017.05.022
- Primary Citation of Related Structures:
5UNI - PubMed Abstract:
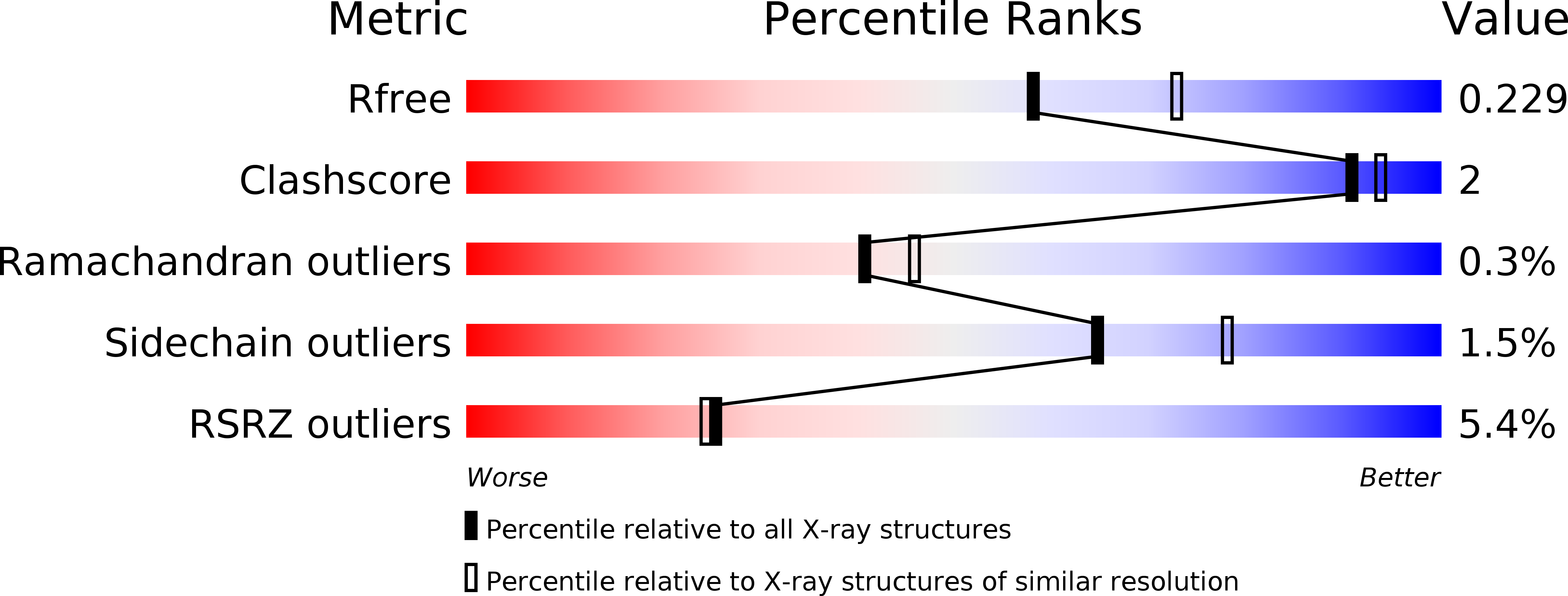
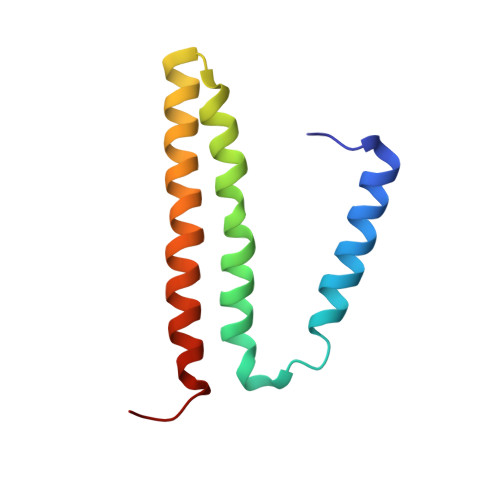
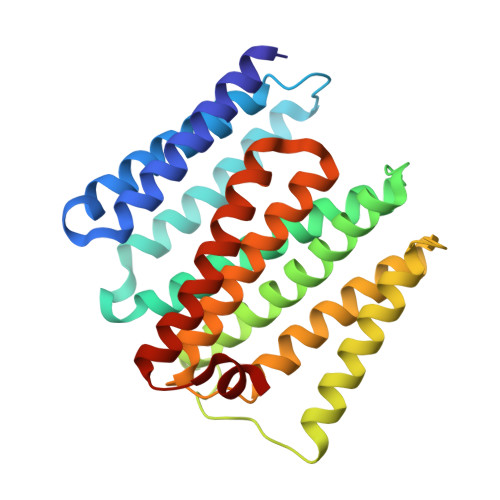
The nicotinamide nucleotide transhydrogenase (TH) is an integral membrane enzyme that uses the proton-motive force to drive hydride transfer from NADH to NADP + in bacteria and eukaryotes. Here we solved a 2.2-Å crystal structure of the TH transmembrane domain (Thermus thermophilus) at pH 6.5. This structure exhibits conformational changes of helix positions from a previous structure solved at pH 8.5, and reveals internal water molecules interacting with residues implicated in proton translocation. Together with molecular dynamics simulations, we show that transient water flows across a narrow pore and a hydrophobic "dry" region in the middle of the membrane channel, with key residues His42 α2 (chain A) being protonated and Thr214 β (chain B) displaying a conformational change, respectively, to gate the channel access to both cytoplasmic and periplasmic chambers. Mutation of Thr214 β to Ala deactivated the enzyme. These data provide new insights into the gating mechanism of proton translocation in TH.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA. Electronic address: piuspada@scripps.edu.