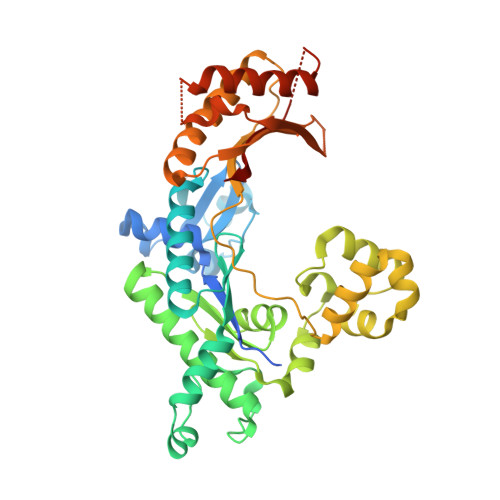
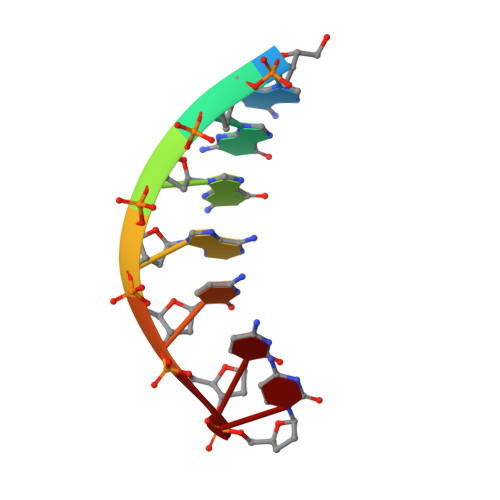
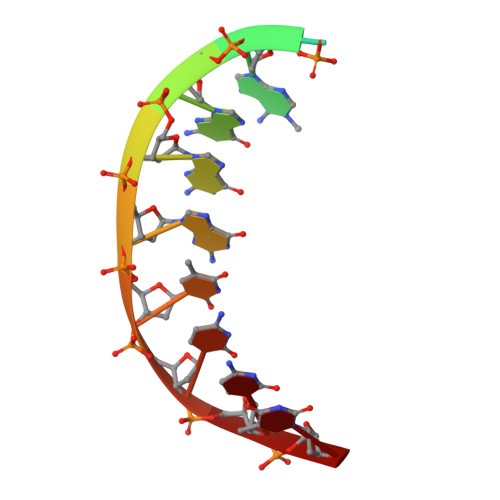
Mechanism of error-free DNA synthesis across N1-methyl-deoxyadenosine by human DNA polymerase-iota.
Jain, R., Choudhury, J.R., Buku, A., Johnson, R.E., Prakash, L., Prakash, S., Aggarwal, A.K.(2017) Sci Rep 7: 43904-43904
- PubMed: 28272441
- DOI: https://doi.org/10.1038/srep43904
- Primary Citation of Related Structures:
5ULW, 5ULX - PubMed Abstract:
N1-methyl-deoxyadenosine (1-MeA) is formed by methylation of deoxyadenosine at the N1 atom. 1-MeA presents a block to replicative DNA polymerases due to its inability to participate in Watson-Crick (W-C) base pairing. Here we determine how human DNA polymerase-ι (Polι) promotes error-free replication across 1-MeA. Steady state kinetic analyses indicate that Polι is ~100 fold more efficient in incorporating the correct nucleotide T versus the incorrect nucleotide C opposite 1-MeA. To understand the basis of this selectivity, we determined ternary structures of Polι bound to template 1-MeA and incoming dTTP or dCTP. In both structures, template 1-MeA rotates to the syn conformation but pairs differently with dTTP versus dCTP. Thus, whereas dTTP partakes in stable Hoogsteen base pairing with 1-MeA, dCTP fails to gain a "foothold" and is largely disordered. Together, our kinetic and structural studies show how Polι maintains discrimination between correct and incorrect incoming nucleotide opposite 1-MeA in preserving genome integrity.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, Box 1677, 1425 Madison Avenue, New York, NY 10029, USA.