Non-canonical reader modules of BAZ1A promote recovery from DNA damage.
Oppikofer, M., Sagolla, M., Haley, B., Zhang, H.M., Kummerfeld, S.K., Sudhamsu, J., Flynn, E.M., Bai, T., Zhang, J., Ciferri, C., Cochran, A.G.(2017) Nat Commun 8: 862-862
- PubMed: 29021563
- DOI: https://doi.org/10.1038/s41467-017-00866-0
- Primary Citation of Related Structures:
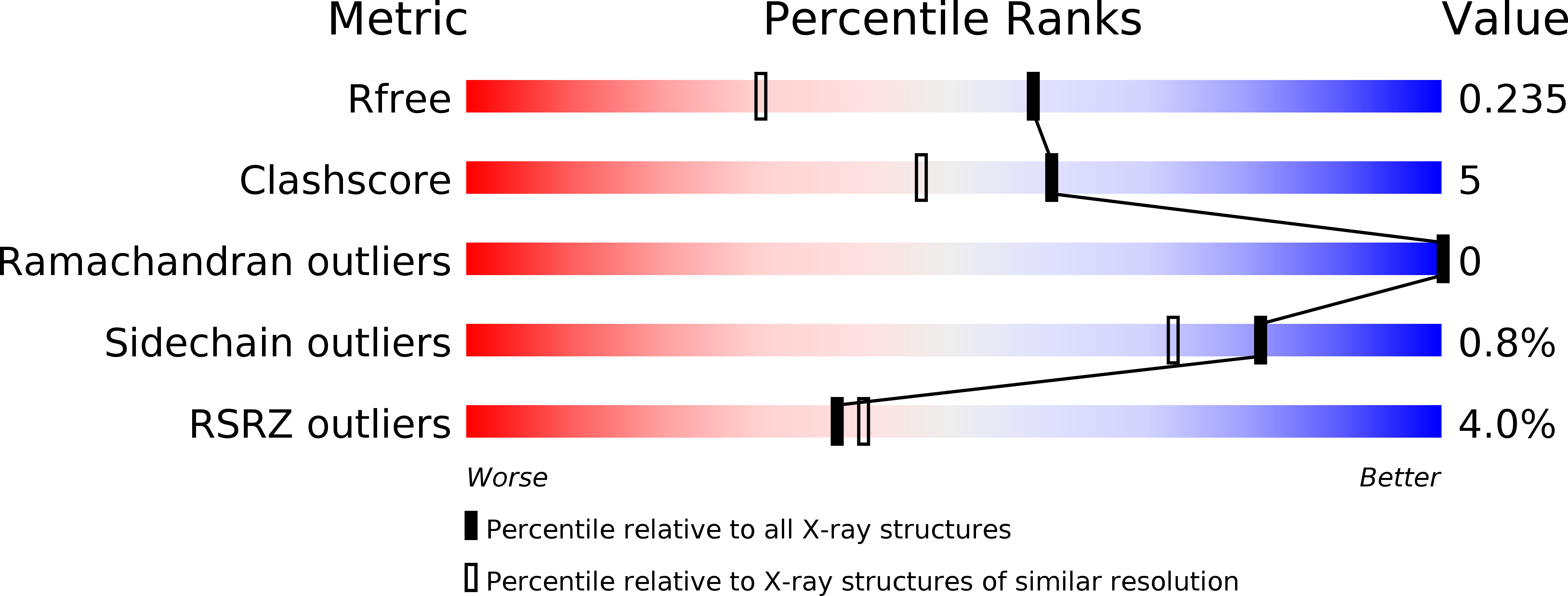
5UIY - PubMed Abstract:
Members of the ISWI family of chromatin remodelers mobilize nucleosomes to control DNA accessibility and, in some cases, are required for recovery from DNA damage. However, it remains poorly understood how the non-catalytic ISWI subunits BAZ1A and BAZ1B might contact chromatin to direct the ATPase SMARCA5. Here, we find that the plant homeodomain of BAZ1A, but not that of BAZ1B, has the unusual function of binding DNA. Furthermore, the BAZ1A bromodomain has a non-canonical gatekeeper residue and binds relatively weakly to acetylated histone peptides. Using CRISPR-Cas9-mediated genome editing we find that BAZ1A and BAZ1B each recruit SMARCA5 to sites of damaged chromatin and promote survival. Genetic engineering of structure-designed bromodomain and plant homeodomain mutants reveals that reader modules of BAZ1A and BAZ1B, even when non-standard, are critical for DNA damage recovery in part by regulating ISWI factors loading at DNA lesions and supporting transcriptional programs required for survival.ISWI chromatin remodelers regulate DNA accessibility and have been implicated in DNA damage repair. Here, the authors uncover functions, in response to DNA damage, for the bromodomain of the ISWI subunit BAZ1B and for the non-canonical PHD and bromodomain modules of the paralog BAZ1A.
Organizational Affiliation:
Department of Early Discovery Biochemistry, Genentech, Inc., 1 DNA Way, South San Francisco, CA, 94080, USA. mariano.oppikofer@pfizer.com.