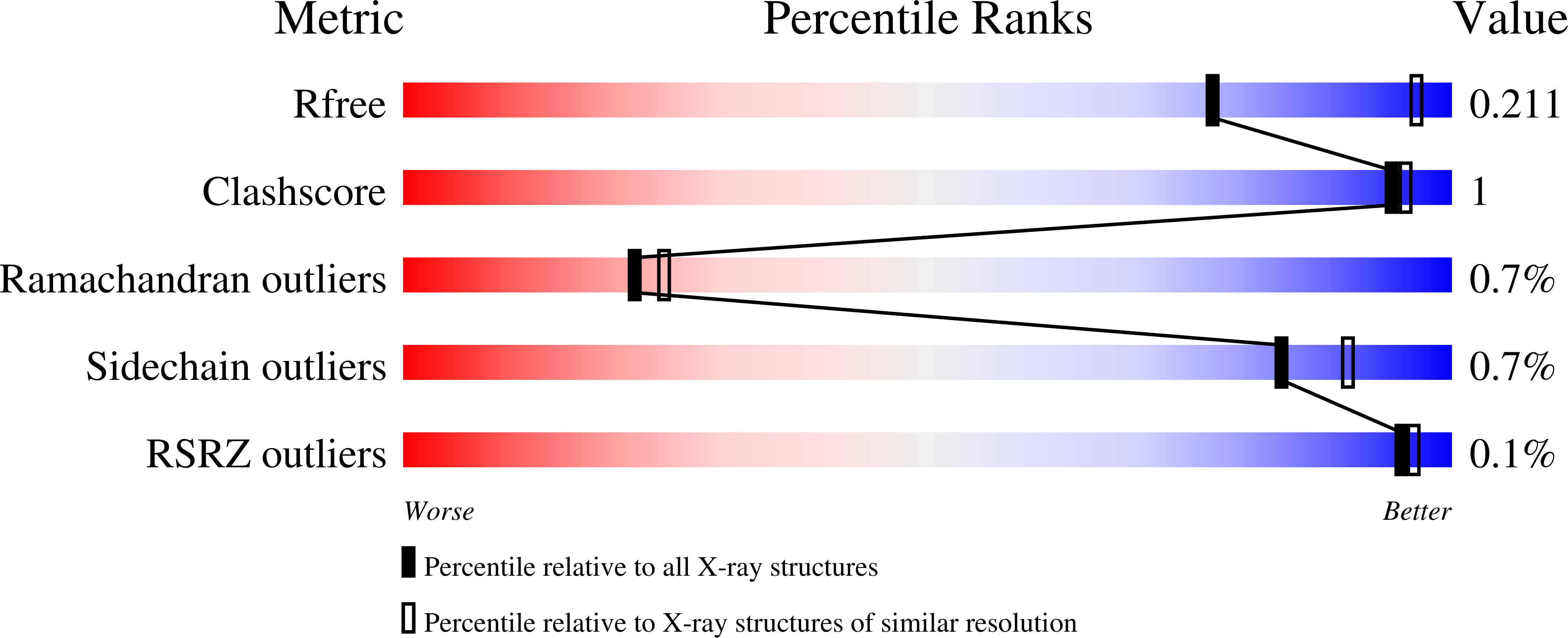
The Role of the Secondary Coordination Sphere in a Fungal Polysaccharide Monooxygenase.
Span, E.A., Suess, D.L.M., Deller, M.C., Britt, R.D., Marletta, M.A.(2017) ACS Chem Biol 12: 1095-1103
- PubMed: 28257189
- DOI: https://doi.org/10.1021/acschembio.7b00016
- Primary Citation of Related Structures:
5UFV - PubMed Abstract:
Polysaccharide monooxygenases (PMOs) are secreted metalloenzymes that catalyze the oxidative degradation of polysaccharides in a copper-, oxygen-, and reductant-dependent manner. Cellulose-active fungal PMOs degrade cellulosic substrates to be utilized as a carbon source for fungal growth. To gain insight into the PMO mechanism, the role of conserved residues in the copper coordination sphere was investigated. Here, we report active-site hydrogen-bonding motifs in the secondary copper coordination sphere of MtPMO3*, a C1-oxidizing PMO from the ascomycete fungus Myceliophthora thermophila. A series of point substitutions that disrupt this conserved network are used to interrogate its function. Activity assays, in conjunction with EPR spectroscopy, demonstrate that residues H161 and Q167 are involved in stabilizing bound oxygen, and H161 appears to play a role in proton transfer. Additionally, Q167 increases the ligand donor strength of Y169 to the copper via a hydrogen-bonding interaction. Altogether, H161 and Q167 are important for oxygen activation, and the results are suggestive of a copper-oxyl active intermediate.
Organizational Affiliation:
Biophysics Graduate Group, University of California, Berkeley , Berkeley, California 94720, United States.