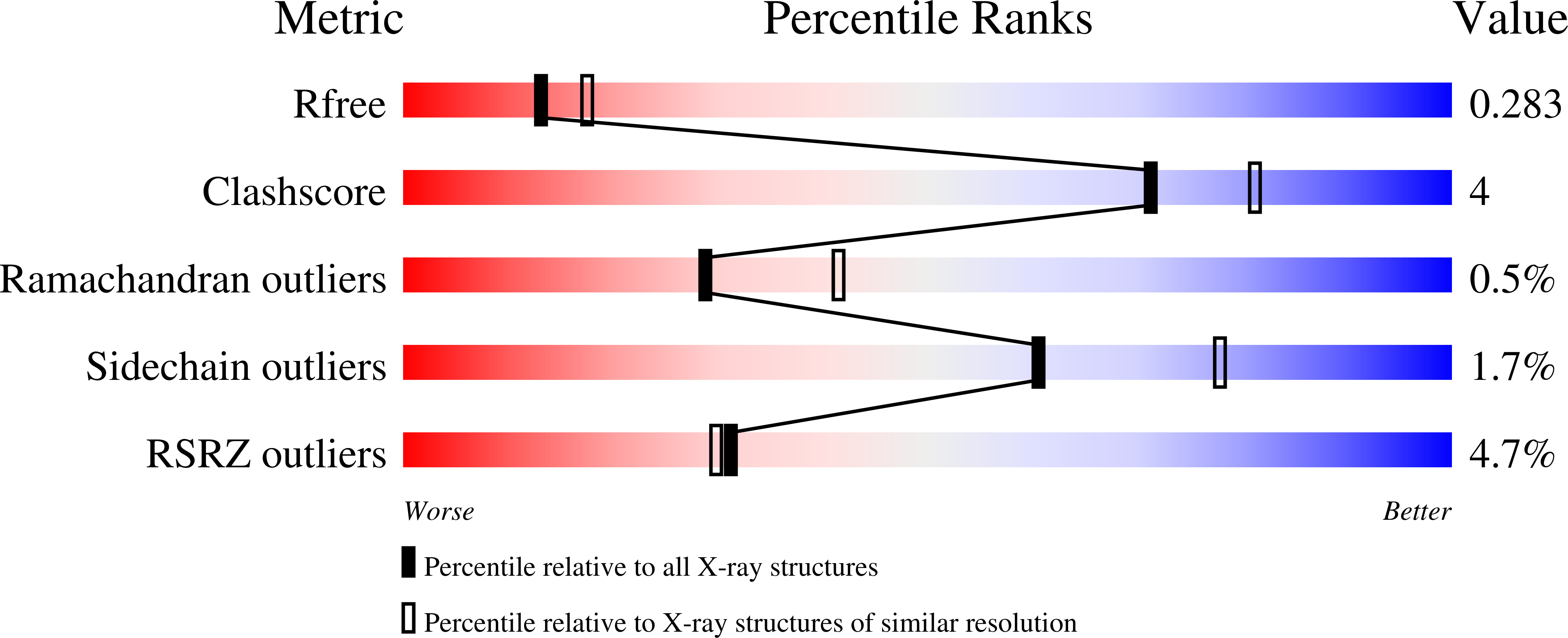
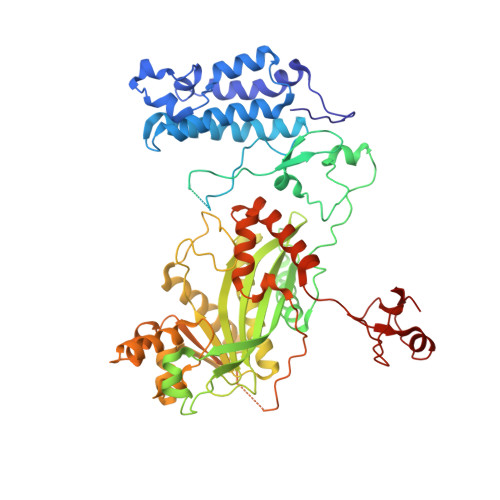
Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation.
Park, S., Stanfield, R.L., Martinez-Yamout, M.A., Dyson, H.J., Wilson, I.A., Wright, P.E.(2017) Proc Natl Acad Sci U S A 114: E5335-E5342
- PubMed: 28630323
- DOI: https://doi.org/10.1073/pnas.1703105114
- Primary Citation of Related Structures:
5U7G - PubMed Abstract:
The histone acetyl transferases CREB-binding protein (CBP) and its paralog p300 play a critical role in numerous cellular processes. Dysregulation of their catalytic activity is associated with several human diseases. Previous work has elucidated the regulatory mechanisms of p300 acetyltransferase activity, but it is not known whether CBP activity is controlled similarly. Here, we present the crystal structure of the CBP catalytic core encompassing the bromodomain (BRD), CH2 (comprising PHD and RING), HAT, and ZZ domains at 2.4-Å resolution. The BRD, PHD, and HAT domains form an integral structural unit to which the RING and ZZ domains are flexibly attached. The structure of the apo-CBP HAT domain is similar to that of acyl-CoA-bound p300 HAT complexes and shows that the acetyl-CoA binding site is stably formed in the absence of cofactor. The BRD, PHD, and ZZ domains interact with small ubiquitin-like modifier 1 (SUMO-1) and Ubc9, and function as an intramolecular E3 ligase for SUMOylation of the cell cycle regulatory domain 1 (CRD1) of CBP, which is located adjacent to the BRD. In vitro HAT assays suggest that the RING domain, the autoregulatory loop (AL) within the HAT domain, and the ZZ domain do not directly influence catalytic activity, whereas the BRD is essential for histone H3 acetylation in nucleosomal substrates. Several lysine residues in the intrinsically disordered AL are autoacetylated by the HAT domain. Upon autoacetylation, acetyl-K1596 (Ac-K1596) binds intramolecularly to the BRD, competing with histones for binding to the BRD and acting as a negative regulator that inhibits histone H3 acetylation.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037.