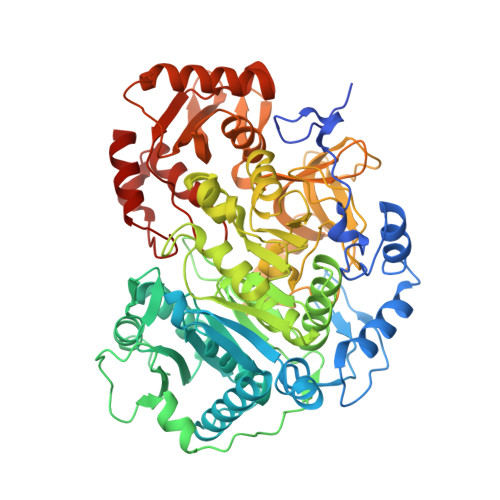
Crystal structure of Cryptococcus neoformans H99 Acetyl-CoA Synthetase in complex with Ac-AMS
Seattle Structural Genomics Center for Infectious Disease (SSGCID), SSGCID, Fox III, D., Potts, K.T., Taylor, B.M., Edwards, T.E., Lorimer, D.D., Mutz, M.W., Krysan, D.J.To be published.