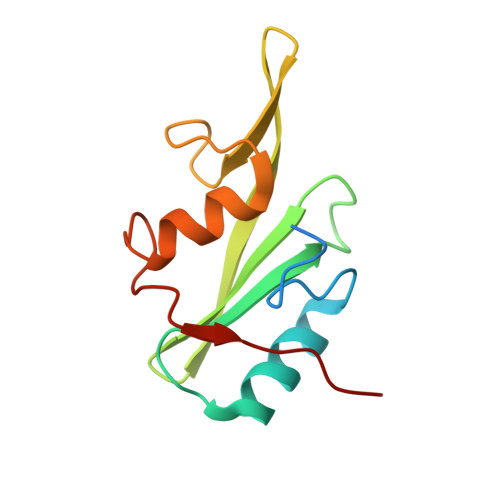
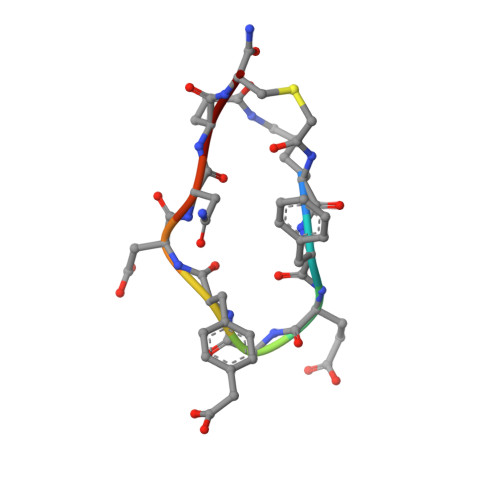
Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
Watson, G.M., Kulkarni, K., Sang, J., Ma, X., Gunzburg, M.J., Perlmutter, P., Wilce, M.C.J., Wilce, J.A.(2017) J Med Chem 60: 9349-9359
- PubMed: 29083893
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01320
- Primary Citation of Related Structures:
5TYI, 5U06, 5U1Q - PubMed Abstract:
Grb7 is a signaling protein with critical roles in tumor cell proliferation and migration and an established cancer therapeutic target. Here we explore chemical space to develop a new bicyclic peptide inhibitor, incorporating thioether and lactam linkers that binds with affinity (K D = 1.1 μM) and specificity to the Grb7-SH2 domain. Structural analysis of the Grb7-SH2/peptide complex revealed an unexpected binding orientation underlying the binding selectivity by this new scaffold. We further incorporated carboxymethylphenylalanine and carboxyphenylalanine phosphotyrosine mimetics and arrived at an optimized inhibitor that potently binds Grb7-SH2 (K D = 0.13 μM) under physiological conditions. X-ray crystal structures of these Grb7-SH2/peptide complexes reveal the structural basis for the most potent and specific inhibitors of Grb7 developed to date. Finally, we demonstrate that cell permeable versions of these peptides successfully block Grb7 mediated interactions in a breast cancer cell line, establishing the potential of these peptides in the development of novel therapeutics targeted to Grb7.
Organizational Affiliation:
Biomedicine Discovery Institute, Department of Biochemistry and Molecular Biology, Monash University , Wellington Road, Clayton, VIC 3800, Australia.