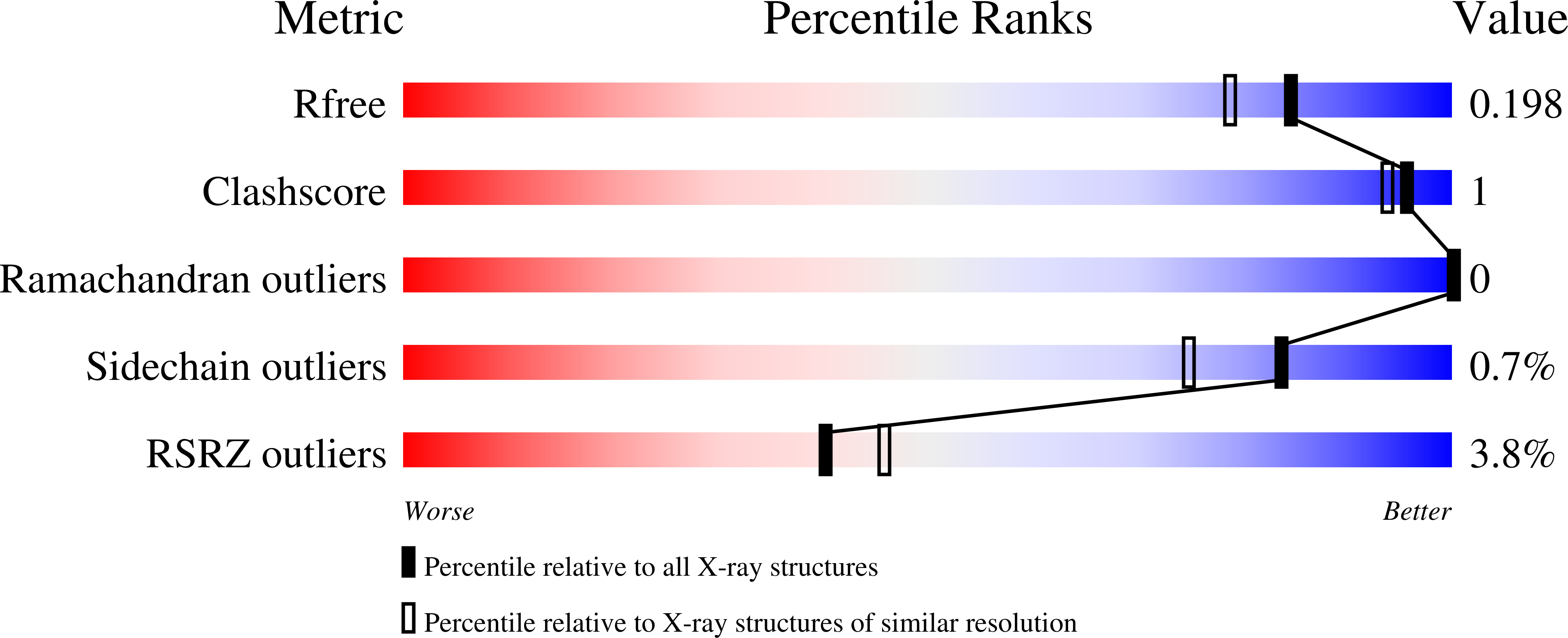
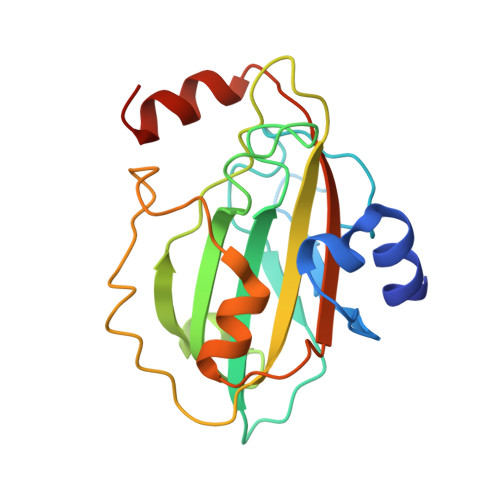
Identification, Characterization, and Structure of Tm16 from Trichuris muris.
Liu, Z., Kelleher, A., Tabb, S., Wei, J., Pollet, J., Hotez, P.J., Bottazzi, M.E., Zhan, B., Asojo, O.A.(2017) J Parasitol Res 2017: 4342789-4342789
- PubMed: 28884022
- DOI: https://doi.org/10.1155/2017/4342789
- Primary Citation of Related Structures:
5TVD - PubMed Abstract:
Trichuriasis is a disease of poverty for which excretory and secretory (ES) products that induce the protective immunity are being investigated as candidate vaccines antigens. In this study, ES products of T. muris and immune sera were produced. The immune sera recognized more than 20 proteins on a 2D-gel of ES products of T. muris adult worms. Tm16 was one of the proteins identified by mass spectrometry. Tm16 shares 57% sequence identity with Ov16, an immunodominant diagnostic antigen from Onchocerca volvulus . Recombinant Tm16 with a carboxyl terminal hexahistidine was produced using Pichia pastoris. Polyclonal antibodies against rTm16 were generated by one-prime and two-boost immunization of three female Balb/c mice with 25 μ g of recombinant Tm16 emulsified with ISA720 adjuvant. These polyclonal antibodies confirmed that Tm16 is localized to the ES products and the soluble fraction of the adult worm. Additionally, the high-resolution crystal structure of Tm16 was solved by molecular replacement. Tm16 belongs to the phosphatidylethanolamine-binding-like protein (PEBP1) family and this is the first structure of a PEBP1 from a parasite.
Organizational Affiliation:
National School of Tropical Medicine, Baylor College of Medicine and Texas Children's Hospital Center for Vaccine Development, Houston TX 77030, USA.