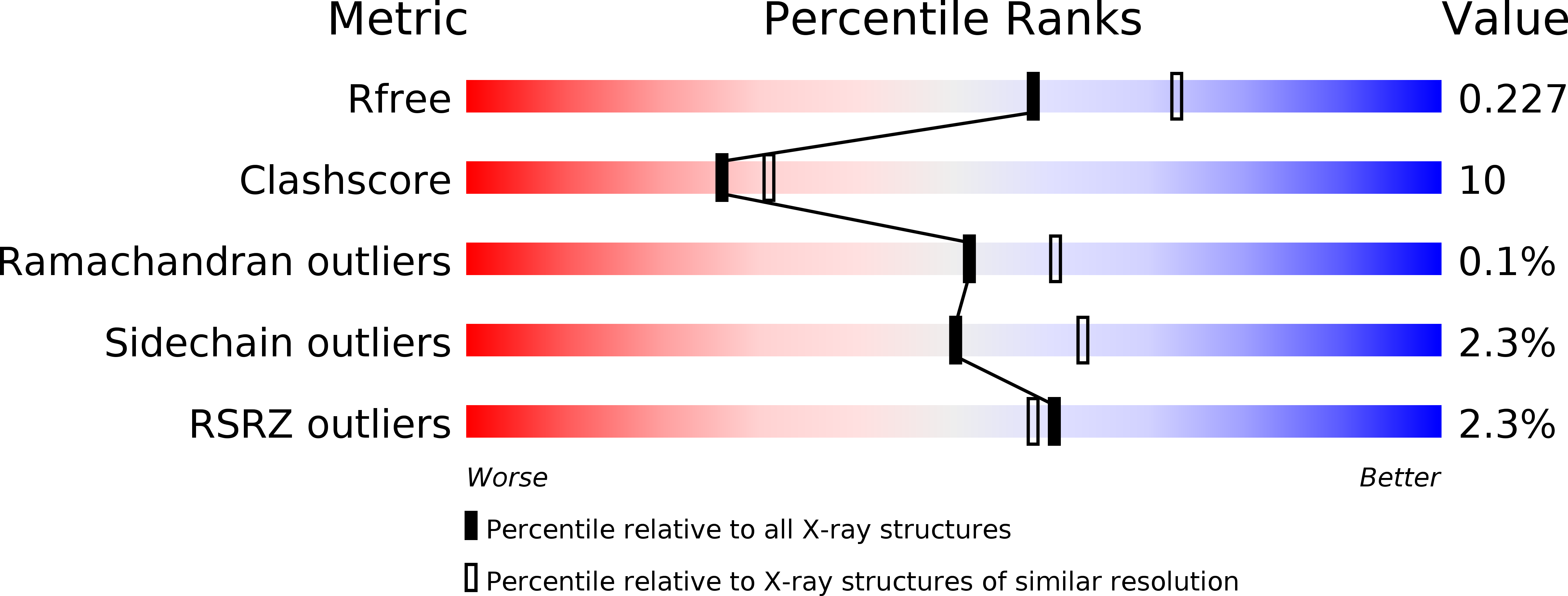
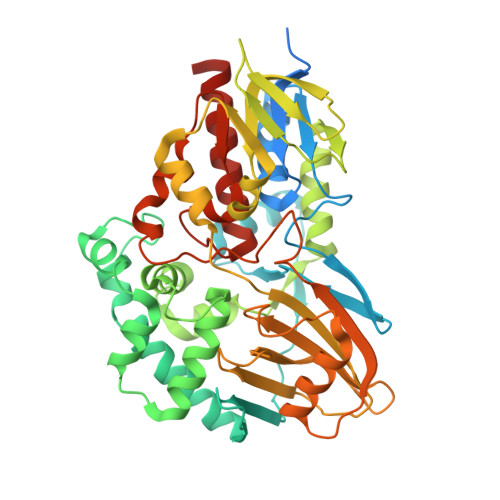
Structural Analysis Provides Mechanistic Insight into Nicotine Oxidoreductase from Pseudomonas putida.
Tararina, M.A., Janda, K.D., Allen, K.N.(2016) Biochemistry 55: 6595-6598
- PubMed: 27933790
- DOI: https://doi.org/10.1021/acs.biochem.6b00963
- Primary Citation of Related Structures:
5TJR, 5TTJ, 5TTK - PubMed Abstract:
The first structure of nicotine oxidoreductase (NicA2) was determined by X-ray crystallography. Pseudomonas putida has evolved nicotine-degrading activity to provide a source of carbon and nitrogen. The structure establishes NicA2 as a member of the monoamine oxidase family. Residues 1-50 are disordered and may play a role in localization. The nicotine-binding site proximal to the isoalloxazine ring of flavin shows an unusual composition of the classical aromatic cage (W427 and N462). The active site architecture is consistent with the proposed binding of the deprotonated form of the substrate and the flavin-dependent oxidation of the pyrrolidone C-N bond followed by nonenzymatic hydrolysis.
Organizational Affiliation:
Program in Biomolecular Pharmacology, Boston University School of Medicine , 72 East Concord Street, Boston, Massachusetts 02118, United States.