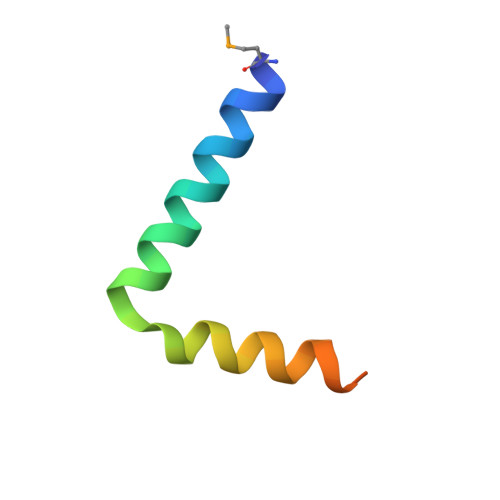
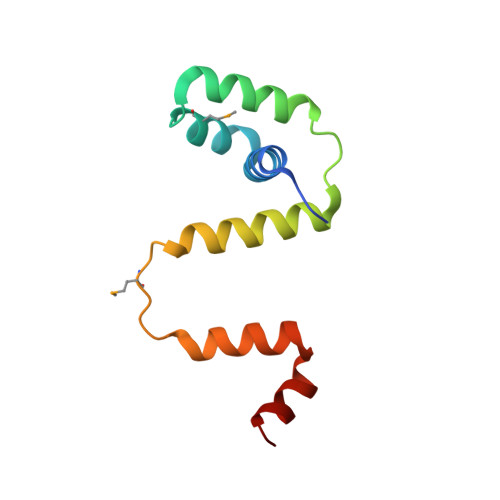
Co-Folding of a FliF-FliG Split Domain Forms the Basis of the MS:C Ring Interface within the Bacterial Flagellar Motor.
Lynch, M.J., Levenson, R., Kim, E.A., Sircar, R., Blair, D.F., Dahlquist, F.W., Crane, B.R.(2017) Structure 25: 317-328
- PubMed: 28089452
- DOI: https://doi.org/10.1016/j.str.2016.12.006
- Primary Citation of Related Structures:
5TDY - PubMed Abstract:
The interface between the membrane (MS) and cytoplasmic (C) rings of the bacterial flagellar motor couples torque generation to rotation within the membrane. The structure of the C-terminal helices of the integral membrane protein FliF (FliF C ) bound to the N terminal domain of the switch complex protein FliG (FliG N ) reveals that FliG N folds around FliF C to produce a topology that closely resembles both the middle and C-terminal domains of FliG. The interface is consistent with solution-state nuclear magnetic resonance, small-angle X-ray scattering, in vivo interaction studies, and cellular motility assays. Co-folding with FliF C induces substantial conformational changes in FliG N and suggests that FliF and FliG have the same stoichiometry within the rotor. Modeling the FliF C :FliG N complex into cryo-electron microscopy rotor density updates the architecture of the middle and upper switch complex and shows how domain shuffling of a conserved interaction module anchors the cytoplasmic rotor to the membrane.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA.