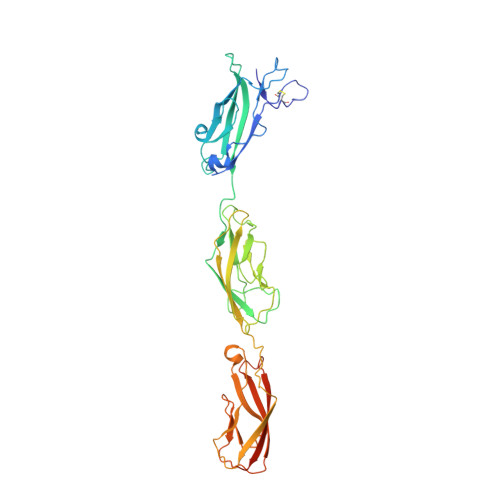
A Partial Calcium-Free Linker Confers Flexibility to Inner-Ear Protocadherin-15.
Powers, R.E., Gaudet, R., Sotomayor, M.(2017) Structure 25: 482-495
- PubMed: 28238533
- DOI: https://doi.org/10.1016/j.str.2017.01.014
- Primary Citation of Related Structures:
5T4M, 5T4N - PubMed Abstract:
Tip links of the inner ear are protein filaments essential for hearing and balance. Two atypical cadherins, cadherin-23 and protocadherin-15, interact in a Ca 2+ -dependent manner to form tip links. The largely unknown structure and mechanics of these proteins are integral to understanding how tip links pull on ion channels to initiate sensory perception. Protocadherin-15 has 11 extracellular cadherin (EC) repeats. Its EC3-4 linker lacks several of the canonical Ca 2+ -binding residues, and contains an aspartate-to-alanine polymorphism (D414A) under positive selection in East Asian populations. We present structures of protocadherin-15 EC3-5 featuring two Ca 2+ -binding linker regions: canonical EC4-5 linker binding three Ca 2+ ions, and non-canonical EC3-4 linker binding only two Ca 2+ ions. Our structures and biochemical assays reveal little difference between the D414 and D414A variants. Simulations predict that the partial Ca 2+ -free EC3-4 linker exhibits increased flexural flexibility without compromised mechanical strength, providing insight into the dynamics of tip links and other atypical cadherins.
Organizational Affiliation:
Department of Molecular and Cellular Biology, Harvard University, 52 Oxford Street, Cambridge, MA 02138, USA; Biophysics Graduate Program, Harvard University, Boston, MA 02115, USA.