Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors.
Clark, K.M., Jenkins, J.L., Fedoriw, N., Dumont, M.E.(2017) Protein Sci 26: 242-257
- PubMed: 27774687
- DOI: https://doi.org/10.1002/pro.3074
- Primary Citation of Related Structures:
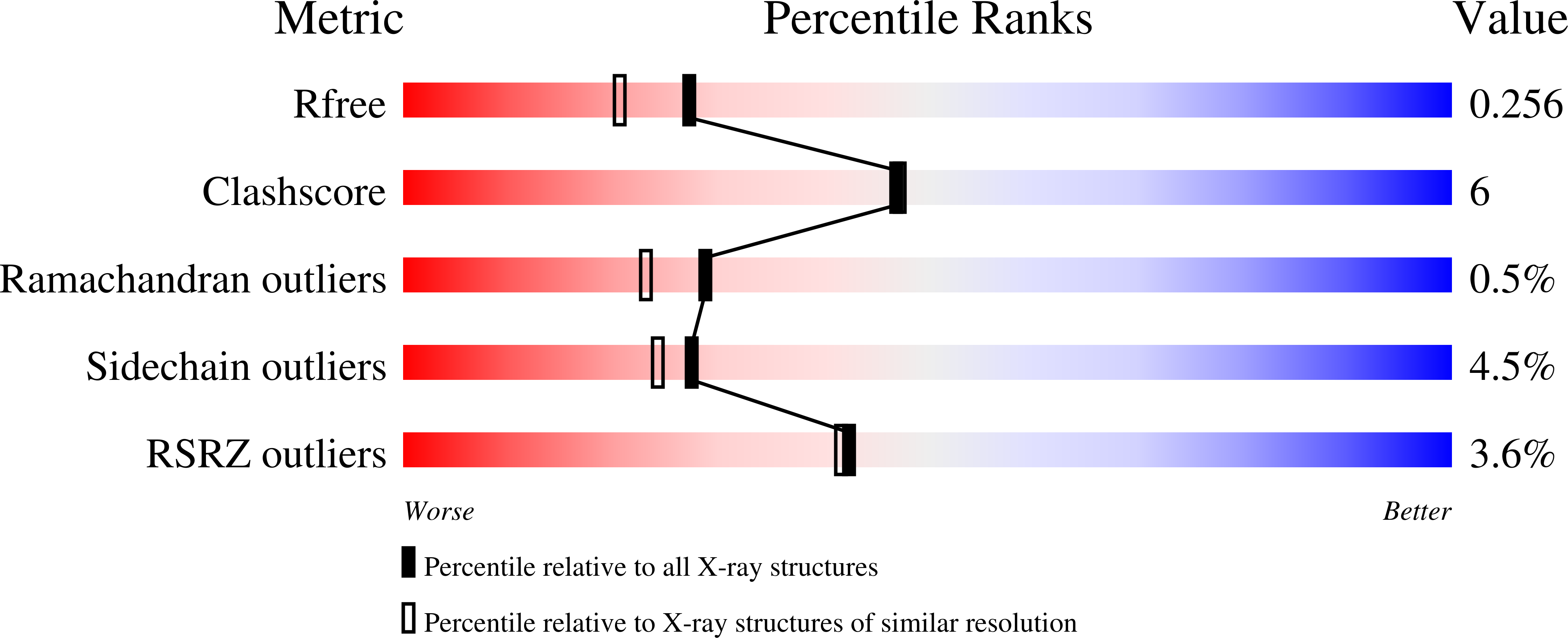
5SYT - PubMed Abstract:
The function and localization of proteins and peptides containing C-terminal "CaaX" (Cys-aliphatic-aliphatic-anything) sequence motifs are modulated by post-translational attachment of isoprenyl groups to the cysteine sulfhydryl, followed by proteolytic cleavage of the aaX amino acids. The zinc metalloprotease ZMPSTE24 is one of two enzymes known to catalyze this cleavage. The only identified target of mammalian ZMPSTE24 is prelamin A, the precursor to the nuclear scaffold protein lamin A. ZMPSTE24 also cleaves prelamin A at a second site 15 residues upstream from the CaaX site. Mutations in ZMPSTE24 result in premature-aging diseases and inhibition of ZMPSTE24 activity has been reported to be an off-target effect of HIV protease inhibitors. We report here the expression (in yeast), purification, and crystallization of human ZMPSTE24 allowing determination of the structure to 2.0 Å resolution. Compared to previous lower resolution structures, the enhanced resolution provides: (1) a detailed view of the active site of ZMPSTE24, including water coordinating the catalytic zinc; (2) enhanced visualization of fenestrations providing access from the exterior to the interior cavity of the protein; (3) a view of the C-terminus extending away from the main body of the protein; (4) localization of ordered lipid and detergent molecules at internal and external surfaces and also projecting through fenestrations; (5) identification of water molecules associated with the surface of the internal cavity. We also used a fluorogenic assay of the activity of purified ZMPSTE24 to demonstrate that HIV protease inhibitors directly inhibit the human enzyme in a manner indicative of a competitive mechanism.
Organizational Affiliation:
Department of Pediatrics, University of Rochester Medical Center, Rochester, New York, 14642.