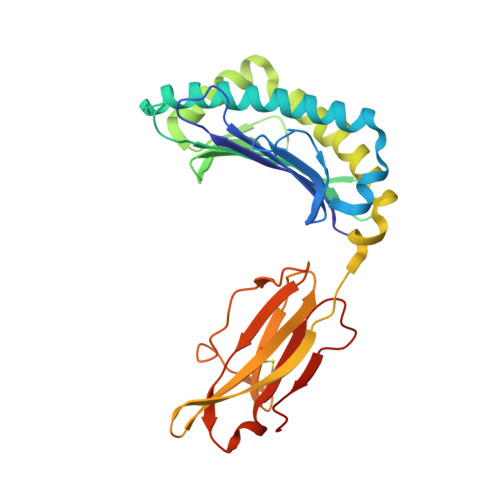
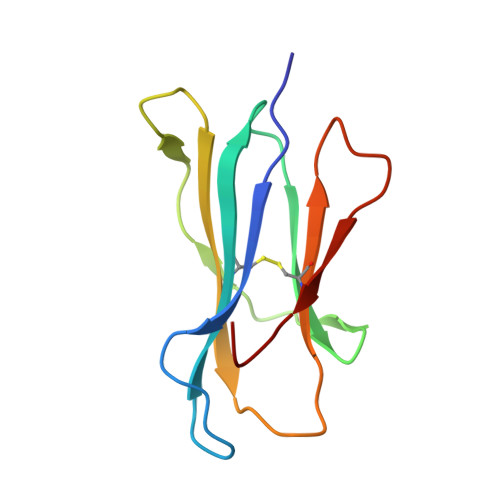
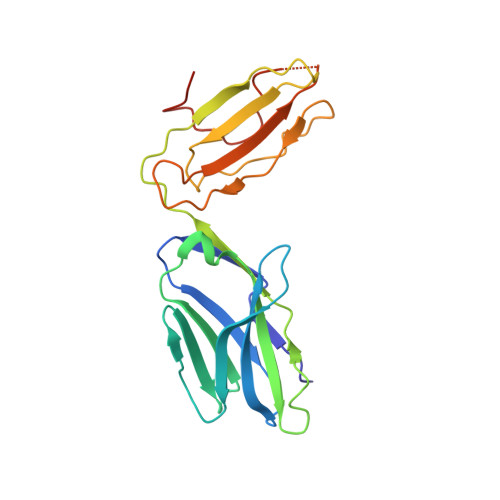
Reversed T Cell Receptor Docking on a Major Histocompatibility Class I Complex Limits Involvement in the Immune Response.
Gras, S., Chadderton, J., Del Campo, C.M., Farenc, C., Wiede, F., Josephs, T.M., Sng, X.Y., Mirams, M., Watson, K.A., Tiganis, T., Quinn, K.M., Rossjohn, J., La Gruta, N.L.(2016) Immunity 45: 749-760
- PubMed: 27717799
- DOI: https://doi.org/10.1016/j.immuni.2016.09.007
- Primary Citation of Related Structures:
5SWS, 5SWZ - PubMed Abstract:
The anti-viral T cell response is drawn from the naive T cell repertoire. During influenza infection, the CD8 + T cell response to an H-2D b -restricted nucleoprotein epitope (NP 366 ) is characterized by preferential expansion of T cells bearing TRBV13 + T cell receptors (TCRs) and avoidance of TRBV17 + T cells, despite the latter dominating the naive precursor repertoire. We found two TRBV17 + TCRs that bound H-2D b -NP 366 with a 180° reversed polarity compared to the canonical TCR-pMHC-I docking. The TRBV17 β-chain dominated the interaction and, whereas the complementarity determining region-3 (CDR3) loops exclusively mediated contacts with the MHC-I, peptide specificity was attributable to germline-encoded recognition. Nevertheless, the TRBV17 + TCR exhibited moderate affinity toward H-2D b -NP 366 and was capable of signal transduction. Thus, the naive CD8 + T cell pool can comprise TCRs adopting reversed pMHC-I docking modes that limit their involvement in the immune response.
Organizational Affiliation:
Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia; ARC Centre of Excellence in Advanced Molecular Imaging, Monash University, Clayton, VIC 3800, Australia.