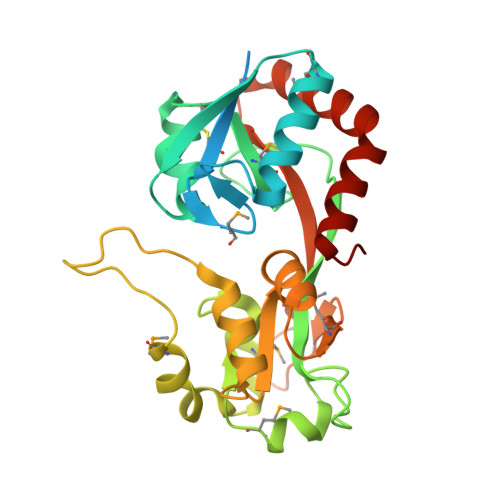
MxaJ structure reveals a periplasmic binding protein-like architecture with unique secondary structural elements
Choi, J.M., Cao, T.P., Kim, S.W., Lee, K.H., Lee, S.H.(2017) Proteins 85: 1379-1386
- PubMed: 28295618
- DOI: https://doi.org/10.1002/prot.25283
- Primary Citation of Related Structures:
5SV6 - PubMed Abstract:
MxaJ is a component of type II methanol dehydrogenase (MDH) that mediates electron transfer during methanol oxidation in methanotrophic bacteria. However, little is known about how MxaJ structurally cooperates with MDH and Cytochrome c L . Here, we report for the first time the crystal structure of MxaJ. MxaJ consists of eight α-helices and six β-strands, and resembles the "bi-lobate" folding architecture found in periplasmic binding proteins. Distinctive features of MxaJ include prominent loops and a β-strand around the hinge region supporting the ligand-binding cavity, which might provide a more favorable framework for interacting with proteins rather than small molecules. Proteins 2017; 85:1379-1386. © 2017 Wiley Periodicals, Inc.
Organizational Affiliation:
Department of Cellular and Molecular Medicine, Chosun University School of Medicine, Gwangju, 61452, Korea.