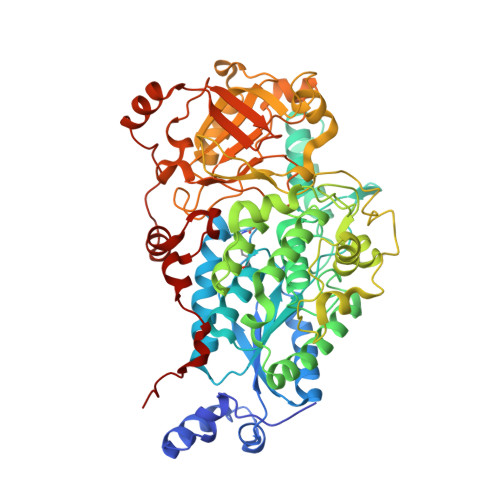
The crystal structure of D-xylonate dehydratase reveals functional features of enzymes from the Ilv/ED dehydratase family.
Rahman, M.M., Andberg, M., Koivula, A., Rouvinen, J., Hakulinen, N.(2018) Sci Rep 8: 865-865
- PubMed: 29339766
- DOI: https://doi.org/10.1038/s41598-018-19192-6
- Primary Citation of Related Structures:
5OYN - PubMed Abstract:
The Ilv/ED dehydratase protein family includes dihydroxy acid-, gluconate-, 6-phosphogluconate- and pentonate dehydratases. The members of this family are involved in various biosynthetic and carbohydrate metabolic pathways. Here, we describe the first crystal structure of D-xylonate dehydratase from Caulobacter crescentus (CcXyDHT) at 2.7 Å resolution and compare it with other available enzyme structures from the IlvD/EDD protein family. The quaternary structure of CcXyDHT is a tetramer, and each monomer is composed of two domains in which the N-terminal domain forms a binding site for a [2Fe-2S] cluster and a Mg 2+ ion. The active site is located at the monomer-monomer interface and contains residues from both the N-terminal recognition helix and the C-terminus of the dimeric counterpart. The active site also contains a conserved Ser490, which probably acts as a base in catalysis. Importantly, the cysteines that participate in the binding and formation of the [2Fe-2S] cluster are not all conserved within the Ilv/ED dehydratase family, which suggests that some members of the IlvD/EDD family may bind different types of [Fe-S] clusters.
Organizational Affiliation:
Department of Chemistry, University of Eastern Finland, PO Box 111, FIN-80101, Joensuu, Finland.