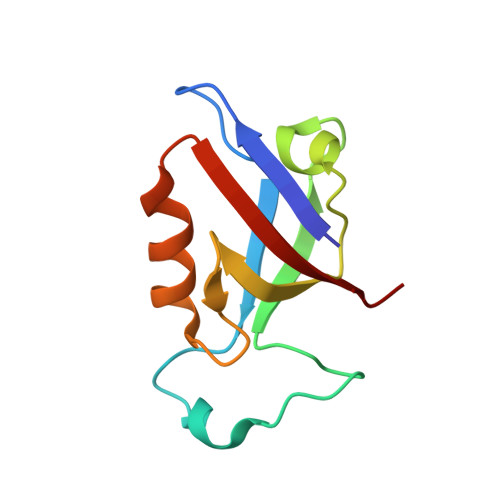
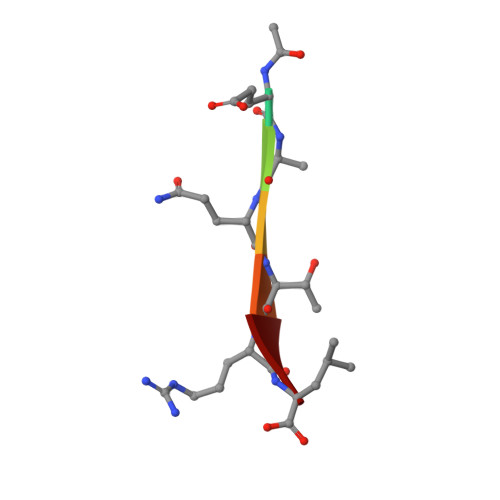
Structural basis for PDZ domain interactions in the post-synaptic density scaffolding protein Shank3.
Ponna, S.K., Ruskamo, S., Myllykoski, M., Keller, C., Boeckers, T.M., Kursula, P.(2018) J Neurochem 145: 449-463
- PubMed: 29473168
- DOI: https://doi.org/10.1111/jnc.14322
- Primary Citation of Related Structures:
5OVA, 5OVC, 5OVP, 5OVV, 6EXJ - PubMed Abstract:
The Shank proteins are crucial scaffolding elements of the post-synaptic density (PSD). One of the best-characterized domains in Shank is the PDZ domain, which binds to C-terminal segments of several other PSD proteins. We carried out a detailed structural analysis of Shank3 PDZ domain-peptide complexes, to understand determinants of binding affinity towards different ligand proteins. Ligand peptides from four different proteins were cocrystallized with the Shank3 PDZ domain, and binding affinities were determined calorimetrically. In addition to conserved class I interactions between the first and third C-terminal peptide residue and Shank3, side chain interactions of other residues in the peptide with the PDZ domain are important factors in defining affinity. Structural conservation suggests that the binding specificities of the PDZ domains from different Shanks are similar. Two conserved buried water molecules in PDZ domains may affect correct local folding of ligand recognition determinants. The solution structure of a tandem Shank3 construct containing the SH3 and PDZ domains showed that the two domains are close to each other, which could be of relevance, when recognizing and binding full target proteins. The SH3 domain did not affect the affinity of the PDZ domain towards short target peptides, and the schizophrenia-linked Shank3 mutation R536W in the linker between the domains had no effect on the structure or peptide interactions of the Shank3 SH3-PDZ unit. Our data show the spatial arrangement of two adjacent Shank domains and pinpoint affinity determinants for short PDZ domain ligands with limited sequence homology.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, Oulu, Finland.