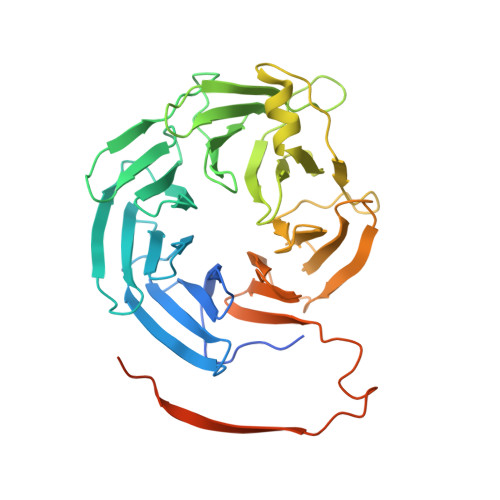
The structure of the RbBP5 beta-propeller domain reveals a surface with potential nucleic acid binding sites.
Mittal, A., Hobor, F., Zhang, Y., Martin, S.R., Gamblin, S.J., Ramos, A., Wilson, J.R.(2018) Nucleic Acids Res 46: 3802-3812
- PubMed: 29897600
- DOI: https://doi.org/10.1093/nar/gky199
- Primary Citation of Related Structures:
5OV3 - PubMed Abstract:
The multi-protein complex WRAD, formed by WDR5, RbBP5, Ash2L and Dpy30, binds to the MLL SET domain to stabilize the catalytically active conformation required for histone H3K4 methylation. In addition, the WRAD complex contributes to the targeting of the activated complex to specific sites on chromatin. RbBP5 is central to MLL catalytic activation, by making critical contacts with the other members of the complex. Interestingly its only major structural domain, a canonical WD40 repeat β-propeller, is not implicated in this function. Here, we present the structure of the RbBP5 β-propeller domain revealing a distinct, feature rich surface, dominated by clusters of Arginine residues. Our nuclear magnetic resonance binding data supports the hypothesis that in addition to the role of RbBP5 in catalytic activation, its β-propeller domain is a platform for the recruitment of the MLL complexes to chromatin targets through its direct interaction with nucleic acids.
Organizational Affiliation:
The Francis Crick Institute, 1 Midland Road, London NW1 1AT, UK.