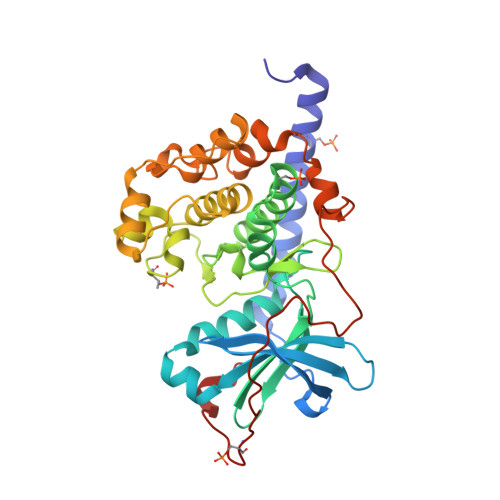
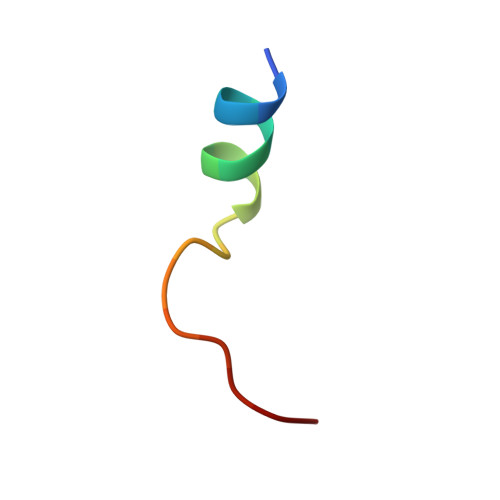
Diamondoid Amino Acid-Based Peptide Kinase A Inhibitor Analogues.
Muller, J., Kirschner, R.A., Berndt, J.P., Wulsdorf, T., Metz, A., Hrdina, R., Schreiner, P.R., Geyer, A., Klebe, G.(2019) ChemMedChem 14: 663-672
- PubMed: 30677243
- DOI: https://doi.org/10.1002/cmdc.201800779
- Primary Citation of Related Structures:
5OK3 - PubMed Abstract:
The incorporation of diamondoid amino acids (DAAs) into peptide-like drugs is a general strategy to improve lipophilicity, membrane permeability, and metabolic stability of peptidomimetic pharmaceuticals. We designed and synthesized five novel peptidic DAA-containing kinase inhibitors of protein kinase A using a sophisticated molecular dynamics protocol and solid-phase peptide synthesis. By means of a thermophoresis binding assay, NMR, and crystal structure analysis, we determined the influence of the DAAs on the secondary structure and binding affinity in comparison to the native protein kinase inhibitor, which is purely composed of proteinogenic amino acids. Affinity and binding pose are largely conserved. One variant showed 6.5-fold potency improvement, most likely related to its increased side chain lipophilicity. A second variant exhibited slightly decreased affinity presumably due to loss of hydrogen-bond contacts to surrounding water molecules of the first solvation shell.
Organizational Affiliation:
Institute of Pharmaceutical Chemistry, Philipps-University Marburg, Marbacher Weg 6, 35032, Marburg, Germany.