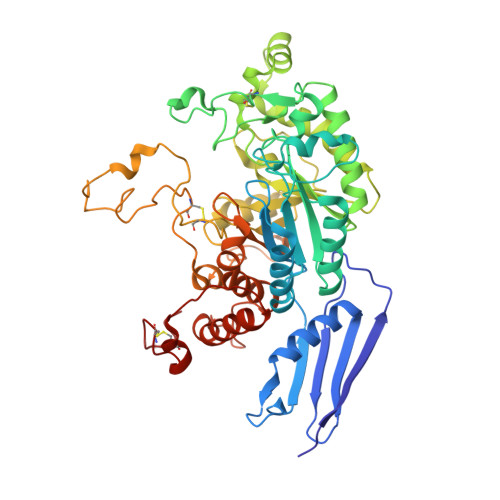
Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae sheds light onto its substrate specificity, high stability, and regulation by propeptide.
Skerlova, J., Blaha, J., Pachl, P., Hofbauerova, K., Kukacka, Z., Man, P., Pompach, P., Novak, P., Otwinowski, Z., Brynda, J., Vanek, O., Rezacova, P.(2018) FEBS J 285: 580-598
- PubMed: 29239122
- DOI: https://doi.org/10.1111/febs.14360
- Primary Citation of Related Structures:
5OAR - PubMed Abstract:
β-N-acetylhexosaminidase from the fungus Aspergillus oryzae is a secreted extracellular enzyme that cleaves chitobiose into constituent monosaccharides. It belongs to the GH 20 glycoside hydrolase family and consists of two N-glycosylated catalytic cores noncovalently associated with two 10-kDa O-glycosylated propeptides. We used X-ray diffraction and mass spectrometry to determine the structure of A. oryzae β-N-acetylhexosaminidase isolated from its natural source. The three-dimensional structure determined and refined to a resolution of 2.3 Å revealed that this enzyme is active as a uniquely tight dimeric assembly further stabilized by N- and O-glycosylation. The propeptide from one subunit forms extensive noncovalent interactions with the catalytic core of the second subunit in the dimer, and this chain swap suggests the distinctive structural mechanism of the enzyme's activation. Unique structural features of β-N-acetylhexosaminidase from A. oryzae define a very stable and robust framework suitable for biotechnological applications. The crystal structure reported here provides structural insights into the enzyme architecture as well as the detailed configuration of the active site. These insights can be applied to rational enzyme engineering.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry, The Czech Academy of Sciences, Prague, Czech Republic.