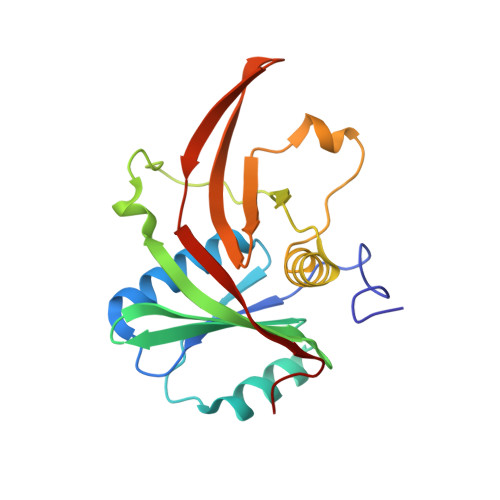
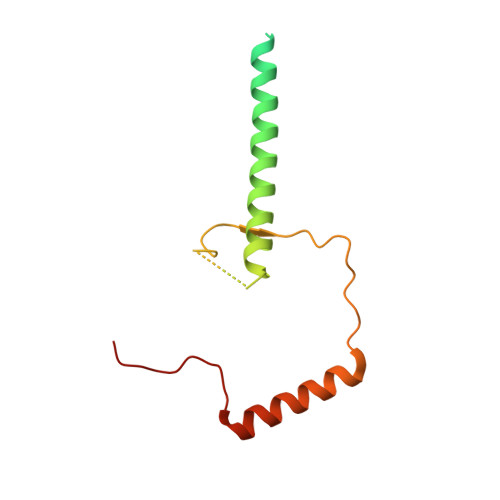
Mpp10 represents a platform for the interaction of multiple factors within the 90S pre-ribosome.
Sa-Moura, B., Kornprobst, M., Kharde, S., Ahmed, Y.L., Stier, G., Kunze, R., Sinning, I., Hurt, E.(2017) PLoS One 12: e0183272-e0183272
- PubMed: 28813493
- DOI: https://doi.org/10.1371/journal.pone.0183272
- Primary Citation of Related Structures:
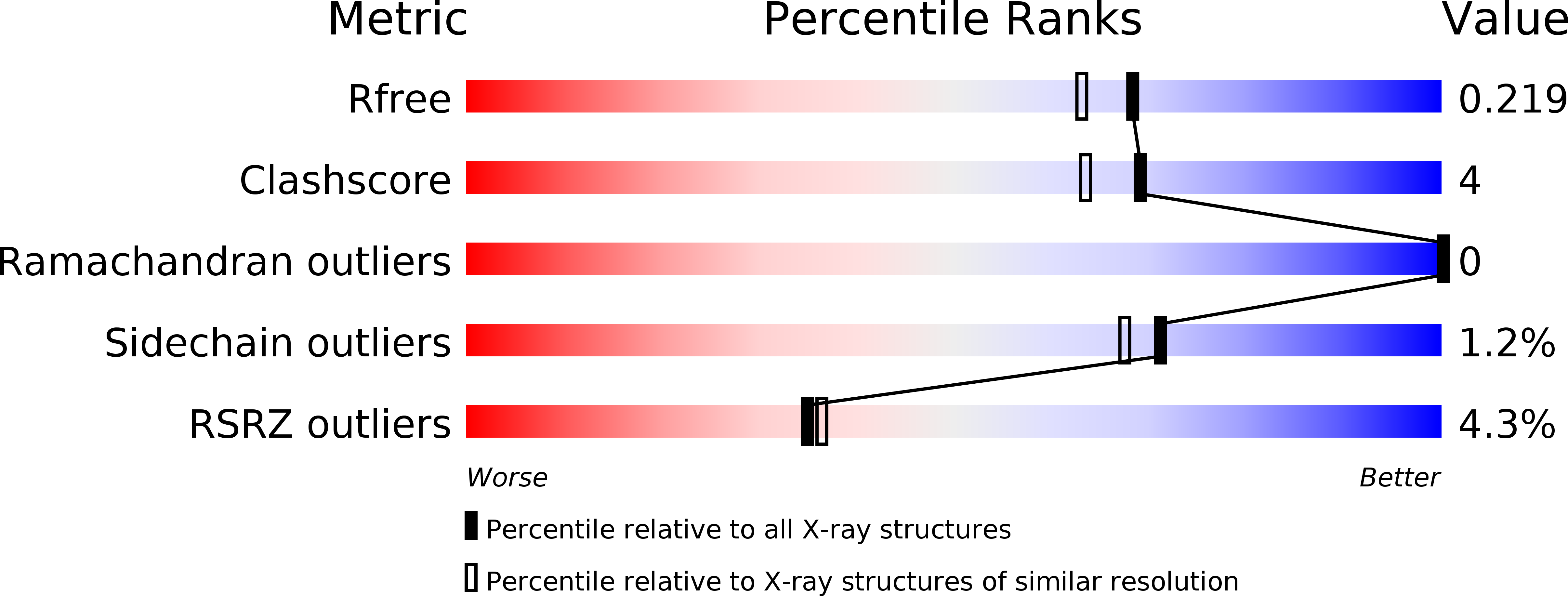
5O9E - PubMed Abstract:
In eukaryotes, ribosome assembly is a highly complex process that involves more than 200 assembly factors that ensure the folding, modification and processing of the different rRNA species as well as the timely association of ribosomal proteins. One of these factors, Mpp10 associates with Imp3 and Imp4 to form a complex that is essential for the normal production of the 18S rRNA. Here we report the crystal structure of a complex between Imp4 and a short helical element of Mpp10 to a resolution of 1.88 Å. Furthermore, we extend the interaction network of Mpp10 and characterize two novel interactions. Mpp10 is able to bind the ribosome biogenesis factor Utp3/Sas10 through two conserved motifs in its N-terminal region. In addition, Mpp10 interacts with the ribosomal protein S5/uS7 using a short stretch within an acidic loop region. Thus, our findings reveal that Mpp10 provides a platform for the simultaneous interaction with multiple proteins in the 90S pre-ribosome.
Organizational Affiliation:
Biochemistry Center Heidelberg BZH, University of Heidelberg, Heidelberg, Germany.