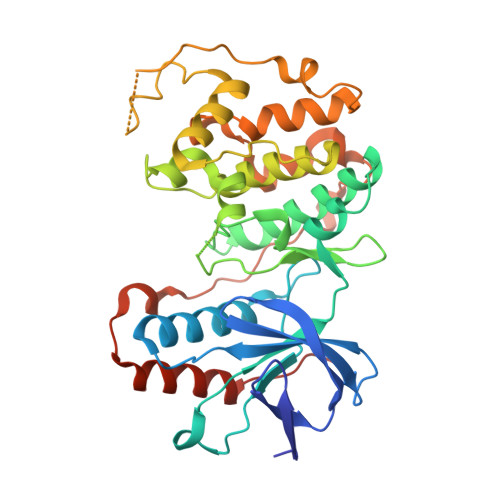
TAB1-Induced Autoactivation of p38 alpha Mitogen-Activated Protein Kinase Is Crucially Dependent on Threonine 185.
Thapa, D., Nichols, C., Bassi, R., Martin, E.D., Verma, S., Conte, M.R., De Santis, V., De Nicola, G.F., Marber, M.S.(2018) Mol Cell Biol 38
- PubMed: 29229647
- DOI: https://doi.org/10.1128/MCB.00409-17
- Primary Citation of Related Structures:
5O90 - PubMed Abstract:
p38α mitogen-activated protein kinase is essential to cellular homeostasis. Two principal mechanisms to activate p38α exist. The first relies on dedicated dual-specificity kinases such as mitogen-activated protein kinase kinase (MAP2K) 3 (MKK3) or 6 (MKK6), which activate p38α by phosphorylating Thr180 and Tyr182 within the activation segment. The second is by autophosphorylation of Thr180 and Tyr182 in cis , mediated by p38α binding the scaffold protein TAB1. The second mechanism occurs during myocardial ischemia, where it aggravates myocardial infarction. Based on the crystal structure of the p38α-TAB1 complex we replaced threonine 185 of p38α with glycine (T185G) to prevent an intramolecular hydrogen bond with Asp150 from being formed. This mutation did not interfere with TAB1 binding to p38α. However, it disrupted the consequent long-range effect of this binding event on the distal activation segment, releasing the constraint on Thr180 that oriented its hydroxyl for phosphotransfer. Based on assays performed in vitro and in vivo , the autoactivation of p38α(T185G) was disabled, while its ability to be activated by upstream MAP2Ks and to phosphorylate downstream substrates remained intact. Furthermore, myocardial cells expressing p38α(T185G) were resistant to injury. These findings reveal a mechanism to selectively disable p38α autoactivation and its consequences, which may ultimately circumvent the toxicity associated with strategies that inhibit p38α kinase activity under all circumstances, such as with ATP-competitive inhibitors.
Organizational Affiliation:
British Heart Foundation Centre of Excellence, Department of Cardiology, The Rayne Institute, St Thomas' Hospital, King's College London, London, United Kingdom.