Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity.
Pao, K.C., Wood, N.T., Knebel, A., Rafie, K., Stanley, M., Mabbitt, P.D., Sundaramoorthy, R., Hofmann, K., van Aalten, D.M.F., Virdee, S.(2018) Nature 556: 381-385
- PubMed: 29643511
- DOI: https://doi.org/10.1038/s41586-018-0026-1
- Primary Citation of Related Structures:
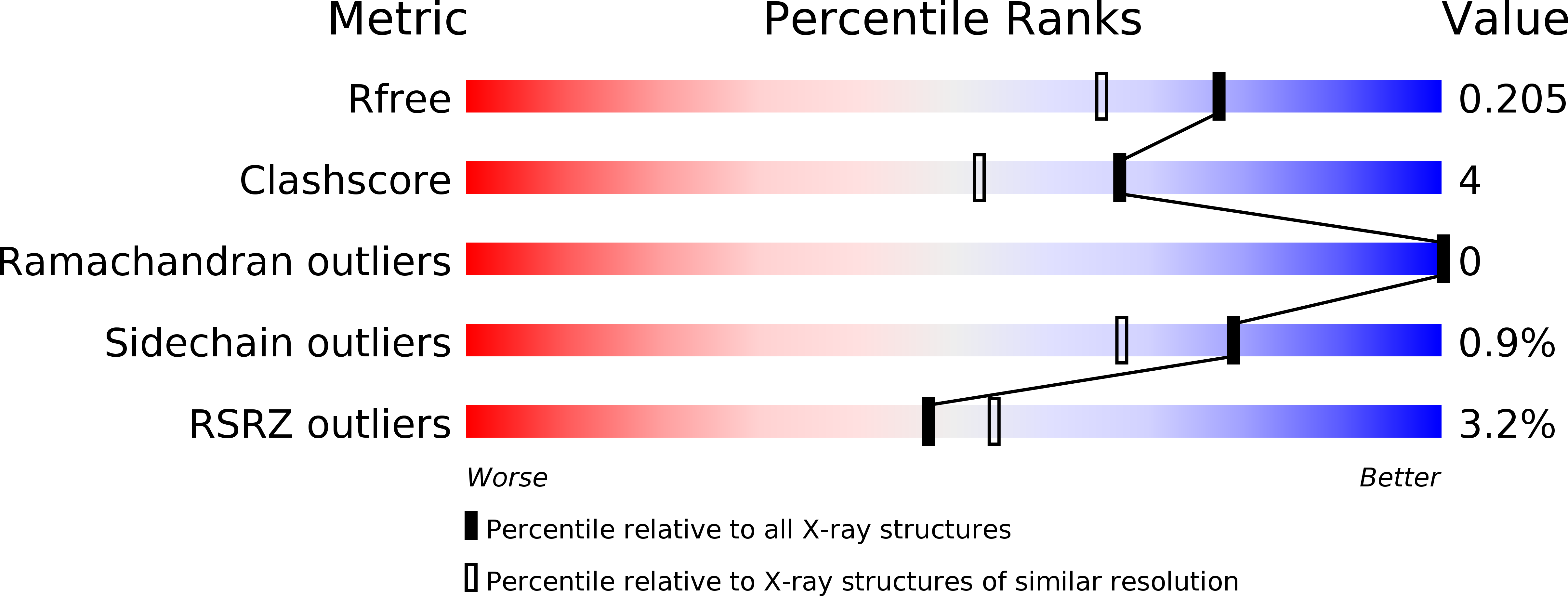
5O6C - PubMed Abstract:
Ubiquitination is initiated by transfer of ubiquitin (Ub) from a ubiquitin-activating enzyme (E1) to a ubiquitin-conjugating enzyme (E2), producing a covalently linked intermediate (E2-Ub) 1 . Ubiquitin ligases (E3s) of the 'really interesting new gene' (RING) class recruit E2-Ub via their RING domain and then mediate direct transfer of ubiquitin to substrates 2 . By contrast, 'homologous to E6-AP carboxy terminus' (HECT) E3 ligases undergo a catalytic cysteine-dependent transthiolation reaction with E2-Ub, forming a covalent E3-Ub intermediate 3,4 . Additionally, RING-between-RING (RBR) E3 ligases have a canonical RING domain that is linked to an ancillary domain. This ancillary domain contains a catalytic cysteine that enables a hybrid RING-HECT mechanism 5 . Ubiquitination is typically considered a post-translational modification of lysine residues, as there are no known human E3 ligases with non-lysine activity. Here we perform activity-based protein profiling of HECT or RBR-like E3 ligases and identify the neuron-associated E3 ligase MYCBP2 (also known as PHR1) as the apparent single member of a class of RING-linked E3 ligase with esterification activity and intrinsic selectivity for threonine over serine. MYCBP2 contains two essential catalytic cysteine residues that relay ubiquitin to its substrate via thioester intermediates. Crystallographic characterization of this class of E3 ligase, which we designate RING-Cys-relay (RCR), provides insights into its mechanism and threonine selectivity. These findings implicate non-lysine ubiquitination in cellular regulation of higher eukaryotes and suggest that E3 enzymes have an unappreciated mechanistic diversity.
Organizational Affiliation:
MRC Protein Phosphorylation and Ubiquitylation Unit, University of Dundee, Dundee, UK.