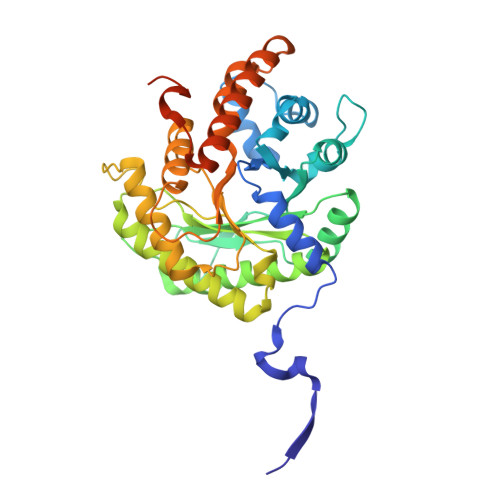
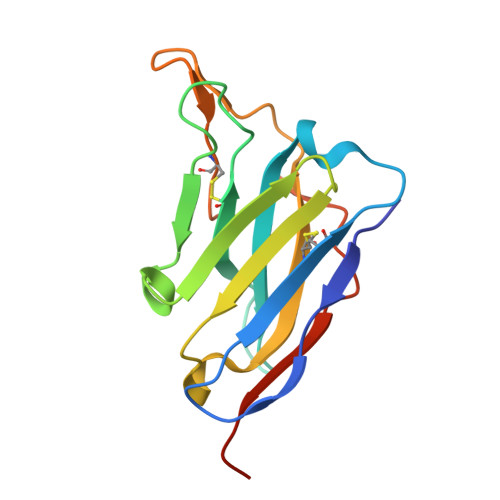
Structural basis for the high specificity of a Trypanosoma congolense immunoassay targeting glycosomal aldolase.
Pinto, J., Odongo, S., Lee, F., Gaspariunaite, V., Muyldermans, S., Magez, S., Sterckx, Y.G.(2017) PLoS Negl Trop Dis 11: e0005932-e0005932
- PubMed: 28915239
- DOI: https://doi.org/10.1371/journal.pntd.0005932
- Primary Citation of Related Structures:
5O0W - PubMed Abstract:
Animal African trypanosomosis (AAT) is a neglected tropical disease which imposes a heavy burden on the livestock industry in Sub-Saharan Africa. Its causative agents are Trypanosoma parasites, with T. congolense and T. vivax being responsible for the majority of the cases. Recently, we identified a Nanobody (Nb474) that was employed to develop a homologous sandwich ELISA targeting T. congolense fructose-1,6-bisphosphate aldolase (TcoALD). Despite the high sequence identity between trypanosomatid aldolases, the Nb474-based immunoassay is highly specific for T. congolense detection. The results presented in this paper yield insights into the molecular principles underlying the assay's high specificity.
Organizational Affiliation:
Research Unit for Cellular and Molecular Immunology (CMIM), Vrije Universiteit Brussel (VUB), Brussels, Belgium.