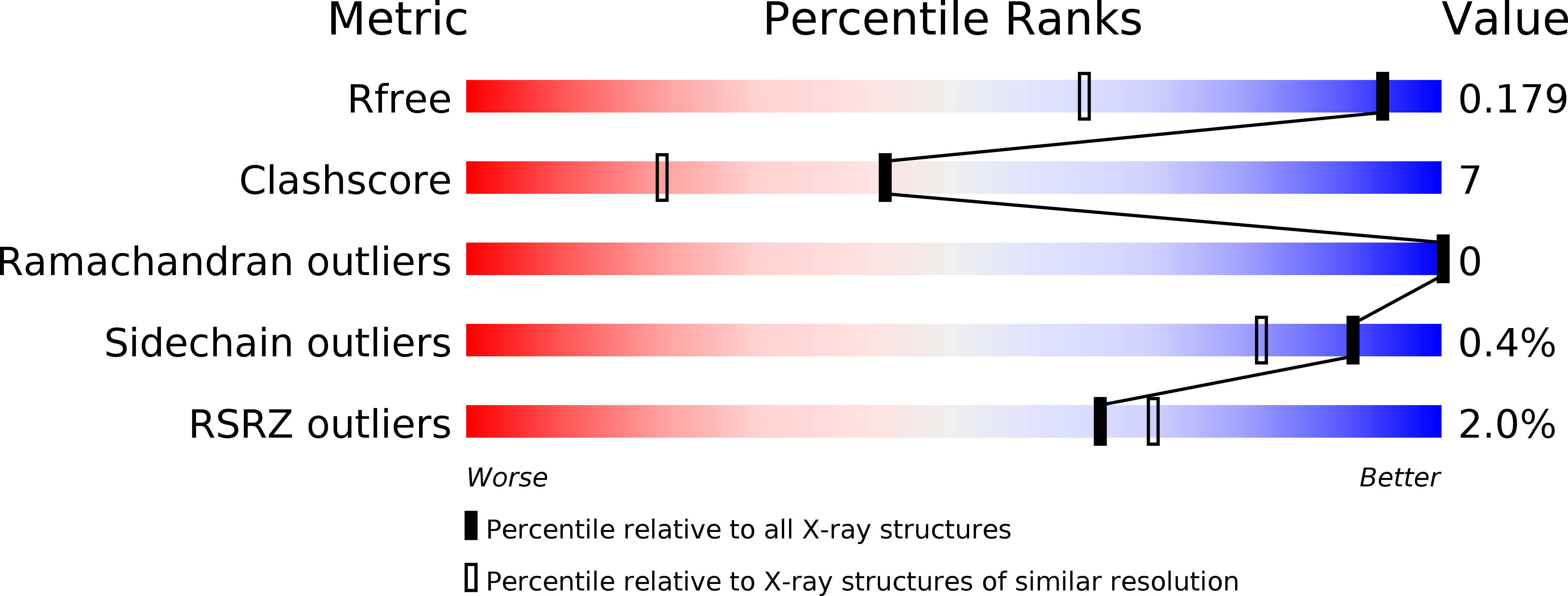
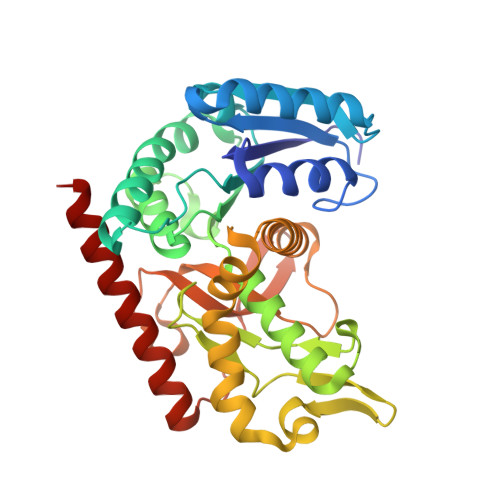
Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
Huang, J., Niazi, A.K., Young, D., Rosado, L.A., Vertommen, D., Bodra, N., Abdelgawwad, M.R., Vignols, F., Wei, B., Wahni, K., Bashandy, T., Bariat, L., Van Breusegem, F., Messens, J., Reichheld, J.P.(2018) J Exp Bot 69: 3491-3505
- PubMed: 29194485
- DOI: https://doi.org/10.1093/jxb/erx396
- Primary Citation of Related Structures:
5NUE, 5NUF - PubMed Abstract:
Plant malate dehydrogenase (MDH) isoforms are found in different cell compartments and function in key metabolic pathways. It is well known that the chloroplastic NADP-dependent MDH activities are strictly redox regulated and controlled by light. However, redox dependence of other NAD-dependent MDH isoforms have been less studied. Here, we show by in vitro biochemical characterization that the major cytosolic MDH isoform (cytMDH1) is sensitive to H2O2 through sulfur oxidation of cysteines and methionines. CytMDH1 oxidation affects the kinetics, secondary structure, and thermodynamic stability of cytMDH1. Moreover, MS analyses and comparison of crystal structures between the reduced and H2O2-treated cytMDH1 further show that thioredoxin-reversible homodimerization of cytMDH1 through Cys330 disulfide formation protects the protein from overoxidation. Consistently, we found that cytosolic thioredoxins interact specifically with cytMDH in a yeast two-hybrid system. Importantly, we also show that cytosolic and chloroplastic, but not mitochondrial NAD-MDH activities are sensitive to H2O2 stress in Arabidopsis. NAD-MDH activities decreased both in a catalase2 mutant and in an NADP-thioredoxin reductase mutant, emphasizing the importance of the thioredoxin-reducing system to protect MDH from oxidation in vivo. We propose that the redox switch of the MDH activity contributes to adapt the cell metabolism to environmental constraints.
Organizational Affiliation:
Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium.