Cross-reactivity between apical membrane antgen 1 and rhoptry neck protein 2 in P. vivax and P. falciparum: A structural and binding study.
Vulliez-Le Normand, B., Saul, F.A., Hoos, S., Faber, B.W., Bentley, G.A.(2017) PLoS One 12: e0183198-e0183198
- PubMed: 28817634
- DOI: https://doi.org/10.1371/journal.pone.0183198
- Primary Citation of Related Structures:
5NQF, 5NQG - PubMed Abstract:
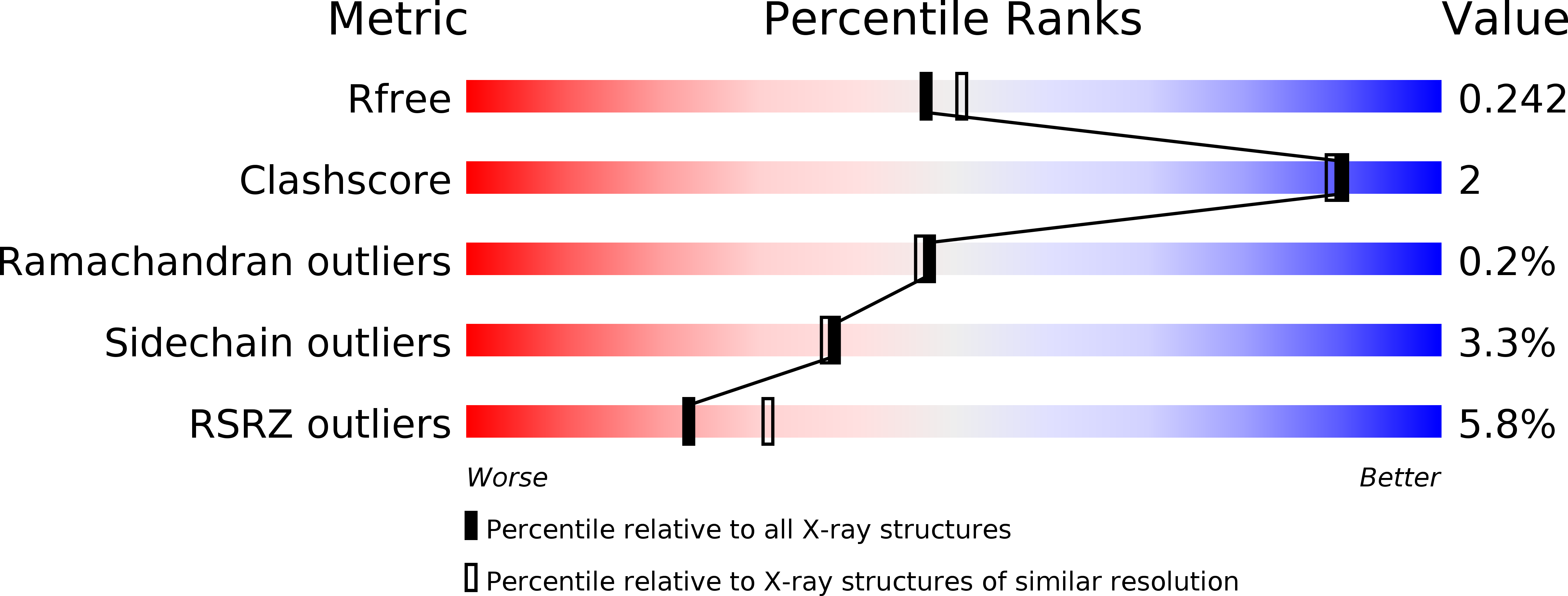
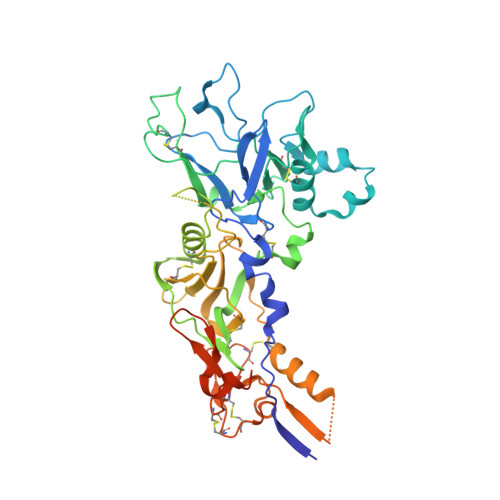
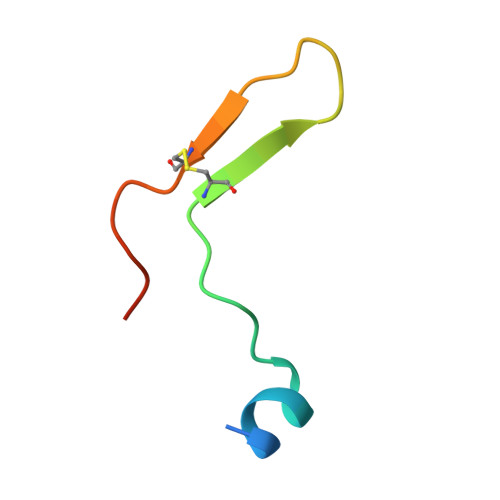
Malaria, a disease endemic in many tropical and subtropical regions, is caused by infection of the erythrocyte by the apicomplexan parasite Plasmodium. Host-cell invasion is a complex process but two Plasmodium proteins, Apical Membrane Antigen 1 (AMA1) and the Rhoptry Neck protein complex (RON), play a key role. AMA1, present on the surface of the parasite, binds tightly to the RON2 component of the RON protein complex, which is inserted into the erythrocyte membrane during invasion. Blocking the AMA1-RON2 interaction with antibodies or peptides inhibits invasion, underlining its importance in the Plasmodium life cycle and as a target for therapeutic strategies. We describe the crystal structure of the complex formed between AMA1 from P. vivax (PvAMA1) and a peptide derived from the externally exposed region of P. vivax RON2 (PvRON2sp1), and of the heterocomplex formed between P. falciparum AMA1 (PfAMA1) and PvRON2sp1. Binding studies show that the affinity of PvRON2sp1 for PvAMA1 is weaker than that previously reported for the PfRON2sp1-PfAMA1 association. Moreover, while PvRON2sp1 shows strong cross-reactivity with PfAMA1, PfRON2sp1 displays no detectable interaction with PvAMA1. The structures show that the equivalent residues PvRON2-Thr2055 and PfRON2-Arg2041 largely account for this pattern of reactivity.
Organizational Affiliation:
Institut Pasteur, Unité de Microbiologie Structurale, Département de Biologie Structurale et Chimie, Centre National de la Recherche Scientifique, UMR 3528, Université Paris Diderot, Sorbonne Paris Cité, Microbiologie Structurale, Paris, France.