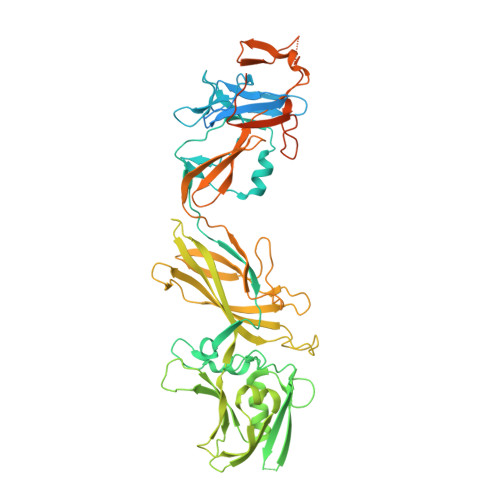
Structural characterization of FlgE2 protein from Helicobacter pylori hook.
Loconte, V., Kekez, I., Matkovic-Calogovic, D., Zanotti, G.(2017) FEBS J 284: 4328-4342
- PubMed: 29083539
- DOI: https://doi.org/10.1111/febs.14312
- Primary Citation of Related Structures:
5NPY - PubMed Abstract:
The Helicobacter pylori flagellum is a complex rotatory nanomachine fundamental for the bacterium's survival in the human stomach. Protein FlgE is a component of the hook, a flexible junction exposed on the cell surface. In the H. pylori genome two different genes are present in different positions coding for hypothetical FlgE. The first protein, FlgE1, is the actual component of the flagellum hook, whilst the second, FlgE2, shares only 26% of the sequence identity with the other and its physiological function is still undefined. We have cloned, purified and crystallized FlgE2, whose structure, determined by the single-wavelength anomalous diffraction method, shows that in overall organization, the protein is composed of three distinct domains, two of them relatively similar to those of FlgE from other Gram-negative bacteria, whilst the third is peculiar to H. pylori. The crystal structure, along with the detected interaction with the regulatory cap protein FlgD, suggests a complementary function of FlgE1 and FlgE2 in the H. pylori flagellum, possibly typical of polar flagella, confirming the role of both proteins in the flagellar hook organization. Although some general features are shared with other Gram-negative bacteria, the presence of two different hook proteins indicates that the molecular organization of H. pylori flagellum has its own peculiarities.
Organizational Affiliation:
Department of Biomedical Sciences, University of Padua, Italy.