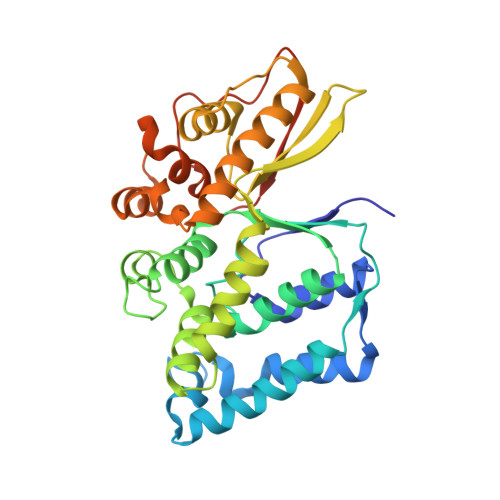
The Legionella effector WipB is a translocated Ser/Thr phosphatase that targets the host lysosomal nutrient sensing machinery.
Prevost, M.S., Pinotsis, N., Dumoux, M., Hayward, R.D., Waksman, G.(2017) Sci Rep 7: 9450-9450
- PubMed: 28842705
- DOI: https://doi.org/10.1038/s41598-017-10249-6
- Primary Citation of Related Structures:
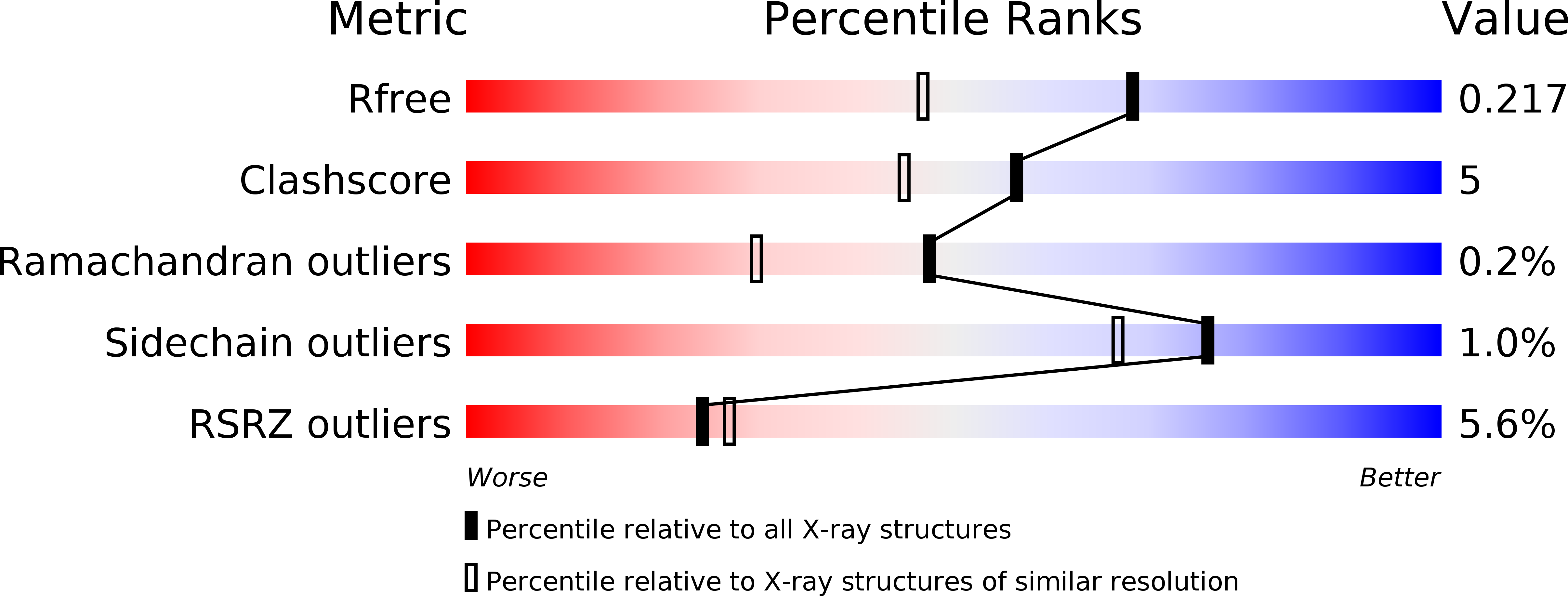
5NNY - PubMed Abstract:
Legionella pneumophila infects human alveolar macrophages and is responsible for Legionnaire's disease, a severe form of pneumonia. L. pneumophila encodes more than 300 putative effectors, which are translocated into the host cell via the Dot/Icm type IV secretion system. These effectors highjack the host's cellular processes to allow bacterial intracellular growth and replication. Here we adopted a multidisciplinary approach to investigate WipB, a Dot/Icm effector of unknown function. The crystal structure of the N-terminal domain at 1.7 Å resolution comprising residues 25 to 344 revealed that WipB harbours a Ser/Thr phosphatase domain related to the eukaryotic phospho-protein phosphatase (PPP) family. The C-terminal domain (residues 365-524) is sufficient to pilot the effector to acidified LAMP1-positive lysosomal compartments, where WipB interacts with the v-ATPase and the associated LAMTOR1 phosphoprotein, key components of the lysosomal nutrient sensing (LYNUS) apparatus that controls the mammalian target of rapamycin (mTORC1) kinase complex at the lysosomal surface. We propose that WipB is a lysosome-targeted phosphatase that modulates cellular nutrient sensing and the control of energy metabolism during Legionella infection.
Organizational Affiliation:
Institute of Structural and Molecular Biology, University College London and Birkbeck, Malet Street, London, WC1E 7HX, UK.