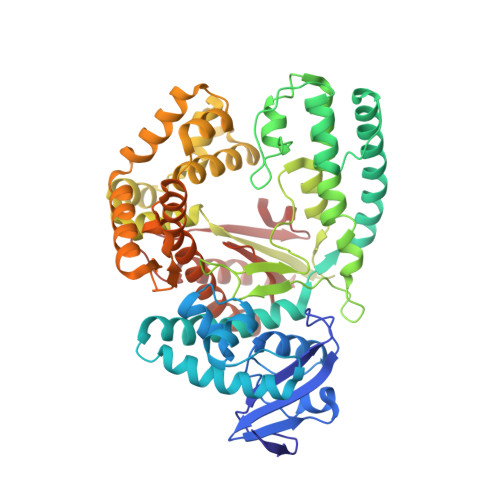
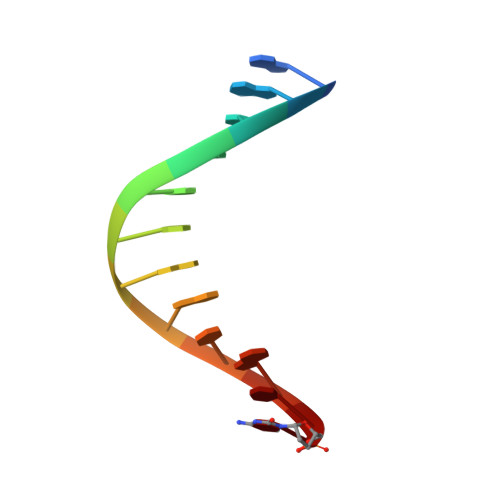
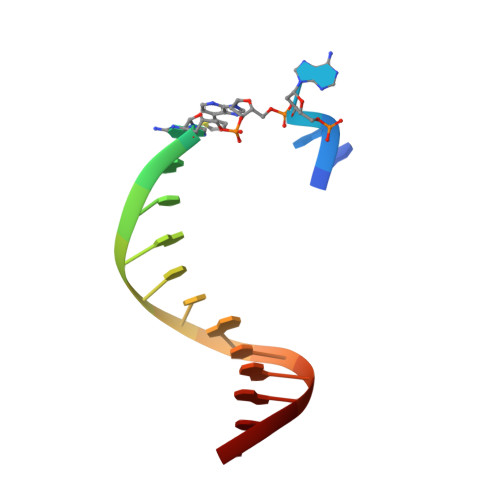
Structural Basis for Expansion of the Genetic Alphabet with an Artificial Nucleobase Pair.
Betz, K., Kimoto, M., Diederichs, K., Hirao, I., Marx, A.(2017) Angew Chem Int Ed Engl 56: 12000-12003
- PubMed: 28594080
- DOI: https://doi.org/10.1002/anie.201704190
- Primary Citation of Related Structures:
5NKL - PubMed Abstract:
Hydrophobic artificial nucleobase pairs without the ability to pair through hydrogen bonds are promising candidates to expand the genetic alphabet. The most successful nucleobase surrogates show little similarity to each other and their natural counterparts. It is thus puzzling how these unnatural molecules are processed by DNA polymerases that have evolved to efficiently work with the natural building blocks. Here, we report structural insight into the insertion of one of the most promising hydrophobic unnatural base pairs, the dDs-dPx pair, into a DNA strand by a DNA polymerase. We solved a crystal structure of KlenTaq DNA polymerase with a modified template/primer duplex bound to the unnatural triphosphate. The ternary complex shows that the artificial pair adopts a planar structure just like a natural nucleobase pair, and identifies features that might hint at the mechanisms accounting for the lower incorporation efficiency observed when processing the unnatural substrates.
Organizational Affiliation:
Departments of Chemistry and Biology and Konstanz Research School Chemical Biology, Universität Konstanz, Universitätsstrasse 10, 78464, Konstanz, Germany.