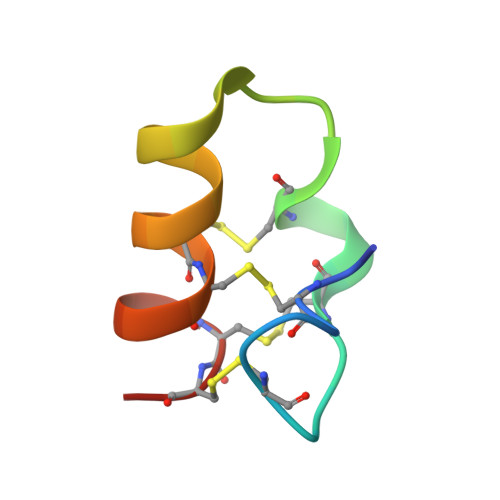
Lybatides from Lycium barbarum Contain An Unusual Cystine-stapled Helical Peptide Scaffold.
Tan, W.L., Wong, K.H., Lei, J., Sakai, N., Tan, H.W., Hilgenfeld, R., Tam, J.P.(2017) Sci Rep 7: 5194-5194
- PubMed: 28701689
- DOI: https://doi.org/10.1038/s41598-017-05037-1
- Primary Citation of Related Structures:
5NGN - PubMed Abstract:
Cysteine-rich peptides (CRPs) of 2-6 kDa are generally thermally and proteolytically stable because of their multiple cross-bracing disulfide bonds. Here, we report the discovery and characterization of two novel cystine-stapled CRPs, designated lybatide 1 and 2 (lyba1 and lyba2), from the cortex of Lycium barbarum root. Lybatides, 32 to 33 amino acids in length, are hyperstable and display a novel disulfide connectivity with a cysteine motif of C-C-C-C-CC-CC which contains two pairs of adjacent cysteines (-CC-CC). X-ray structure analysis revealed the presence of a single cystine-stabilized (α + π)-helix in lyba2, a rare feature of CRPs. Together, our results suggest that lybatides, one of the smallest four-disulfide-constrained plant CRPs, is a new family of CRPs. Additionally, this study provides new insights into the molecular diversity of plant cysteine-rich peptides and the unusual lybatide scaffold could be developed as a useful template for peptide engineering and therapeutic development.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, Singapore, Singapore.