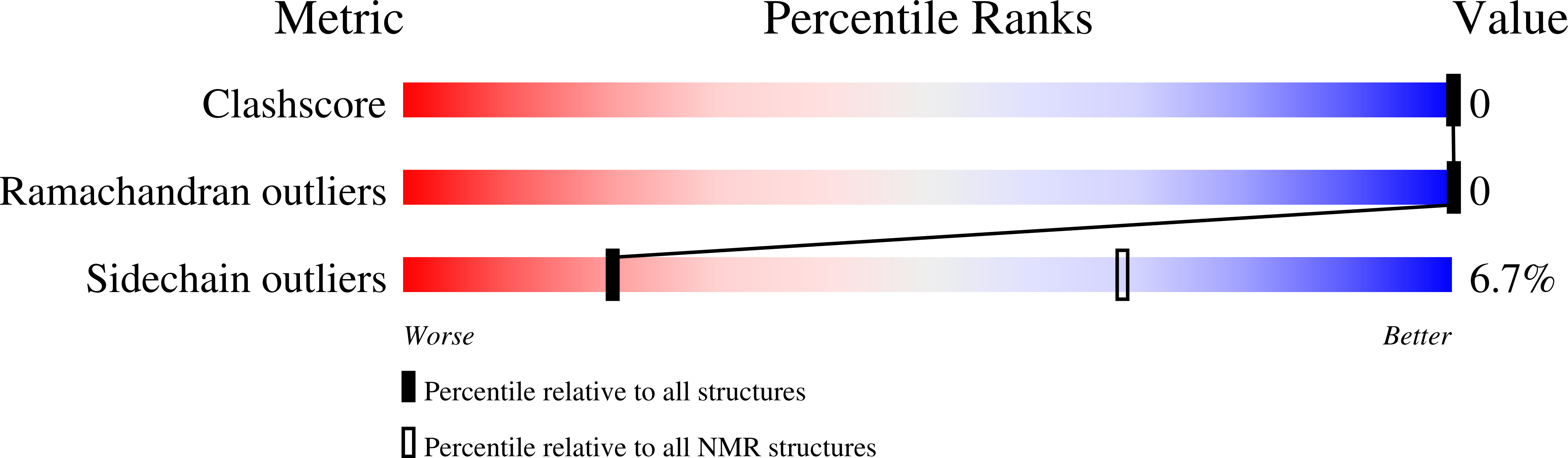Efficient protein production inspired by how spiders make silk.
Kronqvist, N., Sarr, M., Lindqvist, A., Nordling, K., Otikovs, M., Venturi, L., Pioselli, B., Purhonen, P., Landreh, M., Biverstal, H., Toleikis, Z., Sjoberg, L., Robinson, C.V., Pelizzi, N., Jornvall, H., Hebert, H., Jaudzems, K., Curstedt, T., Rising, A., Johansson, J.(2017) Nat Commun 8: 15504-15504
- PubMed: 28534479
- DOI: https://doi.org/10.1038/ncomms15504
- Primary Citation of Related Structures:
5NDA - PubMed Abstract:
Membrane proteins are targets of most available pharmaceuticals, but they are difficult to produce recombinantly, like many other aggregation-prone proteins. Spiders can produce silk proteins at huge concentrations by sequestering their aggregation-prone regions in micellar structures, where the very soluble N-terminal domain (NT) forms the shell. We hypothesize that fusion to NT could similarly solubilize non-spidroin proteins, and design a charge-reversed mutant (NT*) that is pH insensitive, stabilized and hypersoluble compared to wild-type NT. NT*-transmembrane protein fusions yield up to eight times more of soluble protein in Escherichia coli than fusions with several conventional tags. NT* enables transmembrane peptide purification to homogeneity without chromatography and manufacture of low-cost synthetic lung surfactant that works in an animal model of respiratory disease. NT* also allows efficient expression and purification of non-transmembrane proteins, which are otherwise refractory to recombinant production, and offers a new tool for reluctant proteins in general.
Organizational Affiliation:
Division for Neurogeriatrics, Department of NVS, Center for Alzheimer Research, Karolinska Institutet, 141 57 Huddinge, Sweden.