Structural studies of a glycoside hydrolase family 3 beta-glucosidase from the model fungus Neurospora crassa.
Karkehabadi, S., Hansson, H., Mikkelsen, N.E., Kim, S., Kaper, T., Sandgren, M., Gudmundsson, M.(2018) Acta Crystallogr F Struct Biol Commun 74: 787-796
- PubMed: 30511673
- DOI: https://doi.org/10.1107/S2053230X18015662
- Primary Citation of Related Structures:
5NBS - PubMed Abstract:
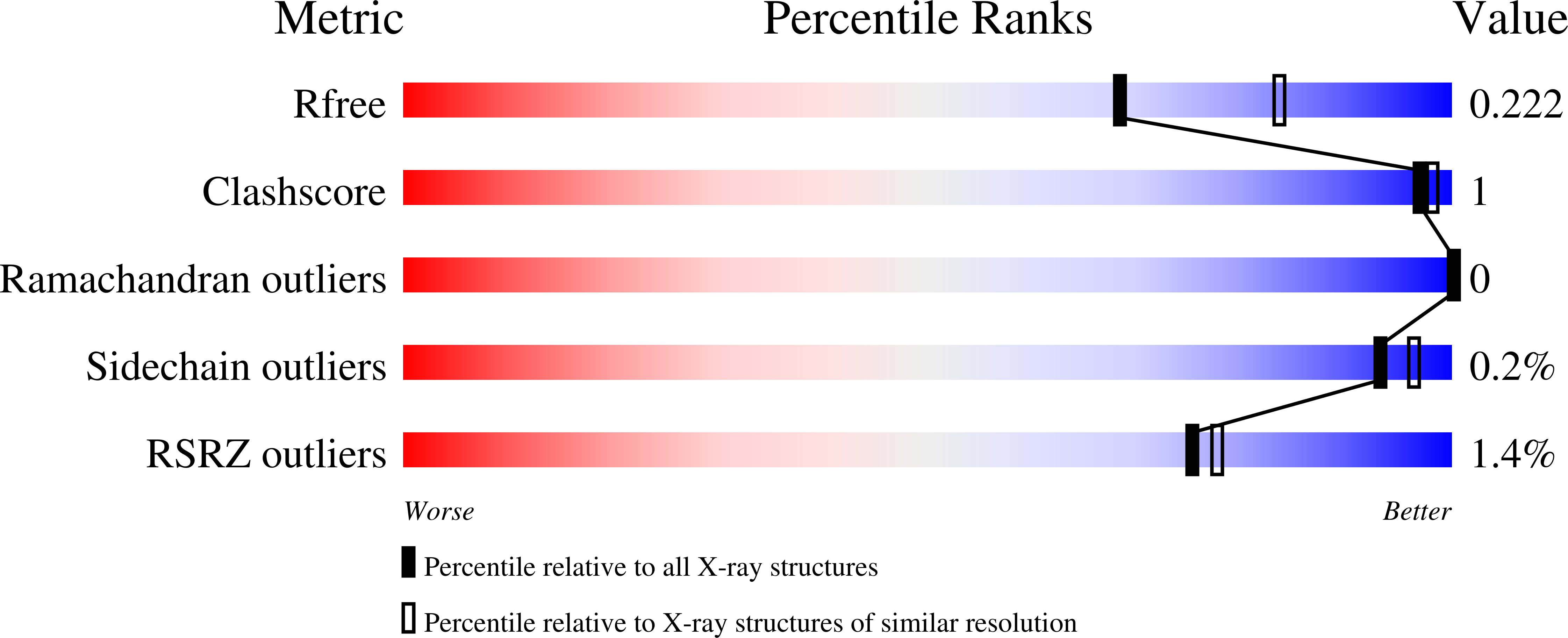
The glycoside hydrolase family 3 (GH3) β-glucosidases are a structurally diverse family of enzymes. Cel3A from Neurospora crassa (NcCel3A) belongs to a subfamily of key enzymes that are crucial for industrial biomass degradation. β-Glucosidases hydrolyse the β-1,4 bond at the nonreducing end of cellodextrins. The hydrolysis of cellobiose is of special importance as its accumulation inhibits other cellulases acting on crystalline cellulose. Here, the crystal structure of the biologically relevant dimeric form of NcCel3A is reported. The structure has been refined to 2.25 Å resolution, with an R cryst and R free of 0.18 and 0.22, respectively. NcCel3A is an extensively N-glycosylated glycoprotein that shares 46% sequence identity with Hypocrea jecorina Cel3A, the structure of which has recently been published, and 61% sequence identity with the thermophilic β-glucosidase from Rasamsonia emersonii. NcCel3A is a three-domain protein with a number of extended loops that deepen the active-site cleft of the enzyme. These structures characterize this subfamily of GH3 β-glucosidases and account for the high cellobiose specificity of this subfamily.
Organizational Affiliation:
Department of Molecular Sciences, Swedish University of Agricultural Sciences, PO Box 7015, SE-750 07 Uppsala, Sweden.