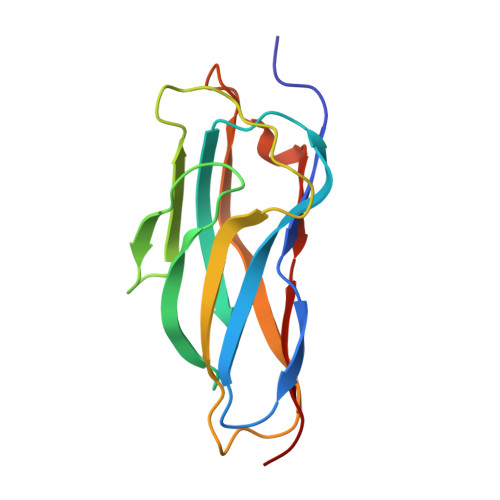
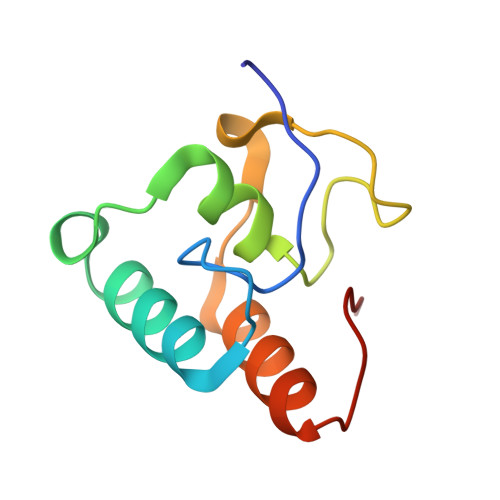
Higher order scaffoldin assembly in Ruminococcus flavefaciens cellulosome is coordinated by a discrete cohesin-dockerin interaction.
Bule, P., Pires, V.M.R., Alves, V.D., Carvalho, A.L., Prates, J.A.M., Ferreira, L.M.A., Smith, S.P., Gilbert, H.J., Noach, I., Bayer, E.A., Najmudin, S., Fontes, C.M.G.A.(2018) Sci Rep 8: 6987-6987
- PubMed: 29725056
- DOI: https://doi.org/10.1038/s41598-018-25171-8
- Primary Citation of Related Structures:
5N5P - PubMed Abstract:
Cellulosomes are highly sophisticated molecular nanomachines that participate in the deconstruction of complex polysaccharides, notably cellulose and hemicellulose. Cellulosomal assembly is orchestrated by the interaction of enzyme-borne dockerin (Doc) modules to tandem cohesin (Coh) modules of a non-catalytic primary scaffoldin. In some cases, as exemplified by the cellulosome of the major cellulolytic ruminal bacterium Ruminococcus flavefaciens, primary scaffoldins bind to adaptor scaffoldins that further interact with the cell surface via anchoring scaffoldins, thereby increasing cellulosome complexity. Here we elucidate the structure of the unique Doc of R. flavefaciens FD-1 primary scaffoldin ScaA, bound to Coh 5 of the adaptor scaffoldin ScaB. The RfCohScaB5-DocScaA complex has an elliptical architecture similar to previously described complexes from a variety of ecological niches. ScaA Doc presents a single-binding mode, analogous to that described for the other two Coh-Doc specificities required for cellulosome assembly in R. flavefaciens. The exclusive reliance on a single-mode of Coh recognition contrasts with the majority of cellulosomes from other bacterial species described to date, where Docs contain two similar Coh-binding interfaces promoting a dual-binding mode. The discrete Coh-Doc interactions observed in ruminal cellulosomes suggest an adaptation to the exquisite properties of the rumen environment.
Organizational Affiliation:
CIISA - Faculdade de Medicina Veterinária, ULisboa, Pólo Universitário do Alto da Ajuda, Avenida da Universidade Técnica, 1300-477, Lisboa, Portugal. pedrobuleg@gmail.com.