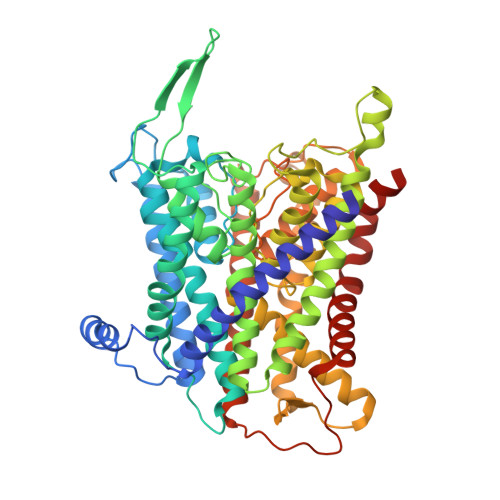
Crystal structure of the potassium-importing KdpFABC membrane complex.
Huang, C.S., Pedersen, B.P., Stokes, D.L.(2017) Nature 546: 681-685
- PubMed: 28636601
- DOI: https://doi.org/10.1038/nature22970
- Primary Citation of Related Structures:
5MRW - PubMed Abstract:
Cellular potassium import systems play a fundamental role in osmoregulation, pH homeostasis and membrane potential in all domains of life. In bacteria, the kdp operon encodes a four-subunit potassium pump that maintains intracellular homeostasis, cell shape and turgor under conditions in which potassium is limiting. This membrane complex, called KdpFABC, has one channel-like subunit (KdpA) belonging to the superfamily of potassium transporters and another pump-like subunit (KdpB) belonging to the superfamily of P-type ATPases. Although there is considerable structural and functional information about members of both superfamilies, the mechanism by which uphill potassium transport through KdpA is coupled with ATP hydrolysis by KdpB remains poorly understood. Here we report the 2.9 Å X-ray structure of the complete Escherichia coli KdpFABC complex with a potassium ion within the selectivity filter of KdpA and a water molecule at a canonical cation site in the transmembrane domain of KdpB. The structure also reveals two structural elements that appear to mediate the coupling between these two subunits. Specifically, a protein-embedded tunnel runs between these potassium and water sites and a helix controlling the cytoplasmic gate of KdpA is linked to the phosphorylation domain of KdpB. On the basis of these observations, we propose a mechanism that repurposes protein channel architecture for active transport across biomembranes.
Organizational Affiliation:
Molecular Biophysics Graduate Program, New York University School of Medicine, Skirball Institute, 540 First Avenue, New York, New York 10016, USA.