Structure of Nora virus at 2.7 angstrom resolution and implications for receptor binding, capsid stability and taxonomy.
Laurinmaki, P., Shakeel, S., Ekstrom, J.O., Mohammadi, P., Hultmark, D., Butcher, S.J.(2020) Sci Rep 10: 19675-19675
- PubMed: 33184473
- DOI: https://doi.org/10.1038/s41598-020-76613-1
- Primary Citation of Related Structures:
5MM2 - PubMed Abstract:
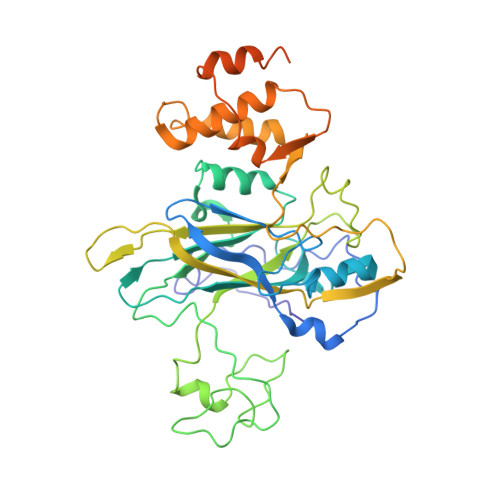
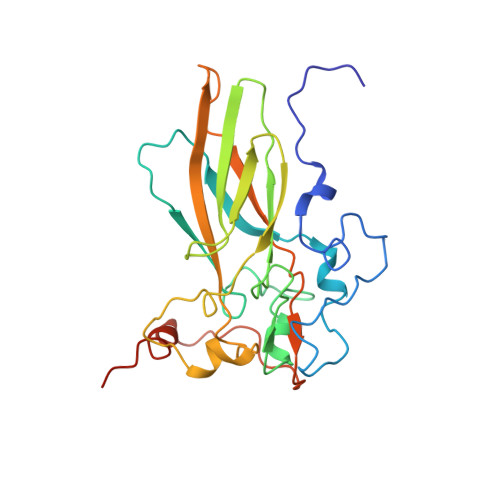
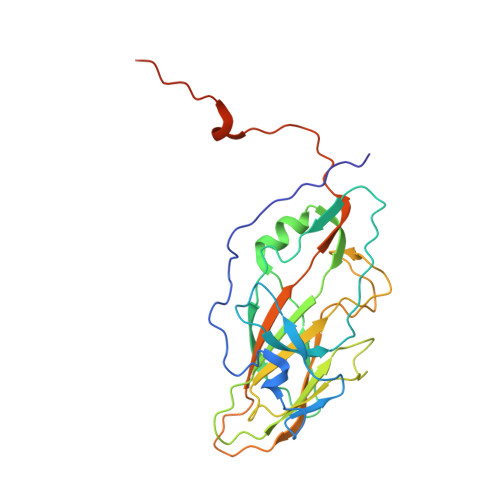
Nora virus, a virus of Drosophila, encapsidates one of the largest single-stranded RNA virus genomes known. Its taxonomic affinity is uncertain as it has a picornavirus-like cassette of enzymes for virus replication, but the capsid structure was at the time for genome publication unknown. By solving the structure of the virus, and through sequence comparison, we clear up this taxonomic ambiguity in the invertebrate RNA virosphere. Despite the lack of detectable similarity in the amino acid sequences, the 2.7 Å resolution cryoEM map showed Nora virus to have T = 1 symmetry with the characteristic capsid protein β-barrels found in all the viruses in the Picornavirales order. Strikingly, α-helical bundles formed from the extended C-termini of capsid protein VP4B and VP4C protrude from the capsid surface. They are similar to signalling molecule folds and implicated in virus entry. Unlike other viruses of Picornavirales, no intra-pentamer stabilizing annulus was seen, instead the intra-pentamer stability comes from the interaction of VP4C and VP4B N-termini. Finally, intertwining of the N-termini of two-fold symmetry-related VP4A capsid proteins and RNA, provides inter-pentamer stability. Based on its distinct structural elements and the genetic distance to other picorna-like viruses we propose that Nora virus, and a small group of related viruses, should have its own family within the order Picornavirales.
Organizational Affiliation:
HiLIFE-Institute of Biotechnology, University of Helsinki, Viikinkaari 9, P.O. Box 56, 00014, Helsinki, Finland.