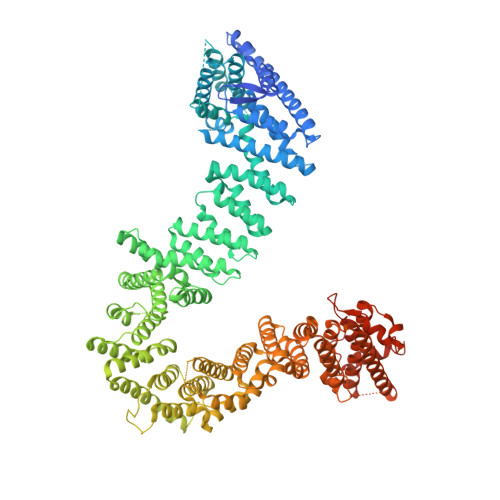
Structure of the cohesin loader Scc2.
Chao, W.C., Murayama, Y., Munoz, S., Jones, A.W., Wade, B.O., Purkiss, A.G., Hu, X.W., Borg, A., Snijders, A.P., Uhlmann, F., Singleton, M.R.(2017) Nat Commun 8: 13952-13952
- PubMed: 28059076
- DOI: https://doi.org/10.1038/ncomms13952
- Primary Citation of Related Structures:
5ME3 - PubMed Abstract:
The functions of cohesin are central to genome integrity, chromosome organization and transcription regulation through its prevention of premature sister-chromatid separation and the formation of DNA loops. The loading of cohesin onto chromatin depends on the Scc2-Scc4 complex; however, little is known about how it stimulates the cohesion-loading activity. Here we determine the large 'hook' structure of Scc2 responsible for catalysing cohesin loading. We identify key Scc2 surfaces that are crucial for cohesin loading in vivo. With the aid of previously determined structures and homology modelling, we derive a pseudo-atomic structure of the full-length Scc2-Scc4 complex. Finally, using recombinantly purified Scc2-Scc4 and cohesin, we performed crosslinking mass spectrometry and interaction assays that suggest Scc2-Scc4 uses its modular structure to make multiple contacts with cohesin.
Organizational Affiliation:
Structural Biology of Chromosome Segregation Laboratory, The Francis Crick Institute, 1 Midland Road, London NW1 1AT, UK.