Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis.
Lasica, A.M., Goulas, T., Mizgalska, D., Zhou, X., de Diego, I., Ksiazek, M., Madej, M., Guo, Y., Guevara, T., Nowak, M., Potempa, B., Goel, A., Sztukowska, M., Prabhakar, A.T., Bzowska, M., Widziolek, M., Thgersen, I.B., Enghild, J.J., Simonian, M., Kulczyk, A.W., Nguyen, K.A., Potempa, J., Gomis-Ruth, F.X.(2016) Sci Rep 6: 37708-37708
- PubMed: 27883039
- DOI: https://doi.org/10.1038/srep37708
- Primary Citation of Related Structures:
5M11 - PubMed Abstract:
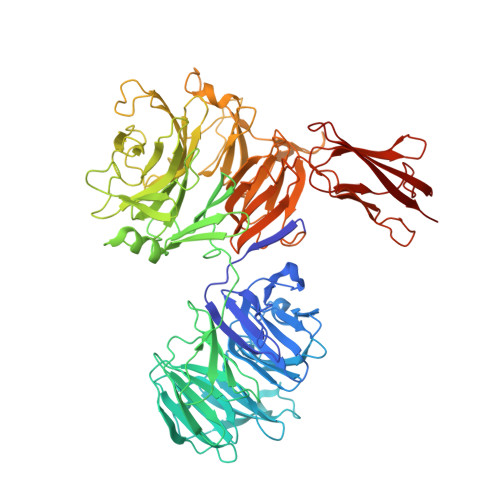
Porphyromonas gingivalis is a member of the human oral microbiome abundant in dysbiosis and implicated in the pathogenesis of periodontal (gum) disease. It employs a newly described type-IX secretion system (T9SS) for secretion of virulence factors. Cargo proteins destined for secretion through T9SS carry a recognition signal in the conserved C-terminal domain (CTD), which is removed by sortase PorU during translocation. Here, we identified a novel component of T9SS, PorZ, which is essential for surface exposure of PorU and posttranslational modification of T9SS cargo proteins. These include maturation of enzyme precursors, CTD removal and attachment of anionic lipopolysaccharide for anchorage in the outer membrane. The crystal structure of PorZ revealed two β-propeller domains and a C-terminal β-sandwich domain, which conforms to the canonical CTD architecture. We further documented that PorZ is itself transported to the cell surface via T9SS as a full-length protein with its CTD intact, independently of the presence or activity of PorU. Taken together, our results shed light on the architecture and possible function of a novel component of the T9SS. Knowledge of how T9SS operates will contribute to our understanding of protein secretion as part of host-microbiome interactions by dysbiotic members of the human oral cavity.
Organizational Affiliation:
Department of Oral Immunology and Infectious Diseases, University of Louisville School of Dentistry, Louisville, KY, USA.