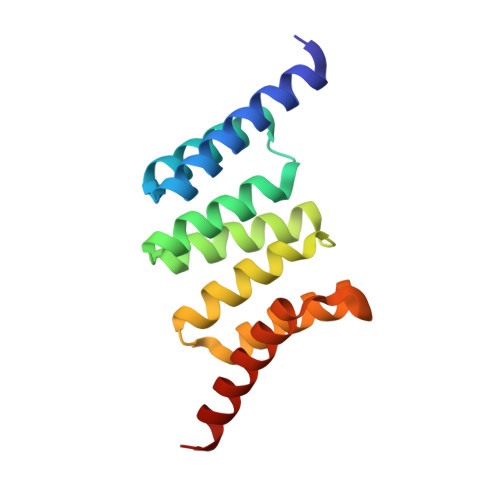
Structure and Interactions of the TPR Domain of Sgt2 with Yeast Chaperones and Ybr137wp.
Krysztofinska, E.M., Evans, N.J., Thapaliya, A., Murray, J.W., Morgan, R.M.L., Martinez-Lumbreras, S., Isaacson, R.L.(2017) Front Mol Biosci 4: 68-68
- PubMed: 29075633
- DOI: https://doi.org/10.3389/fmolb.2017.00068
- Primary Citation of Related Structures:
5LYN, 5LYP - PubMed Abstract:
Small glutamine-rich tetratricopeptide repeat-containing protein 2 (Sgt2) is a multi-module co-chaperone involved in several protein quality control pathways. The TPR domain of Sgt2 and several other proteins, including SGTA, Hop, and CHIP, is a highly conserved motif known to form transient complexes with molecular chaperones such as Hsp70 and Hsp90. In this work, we present the first high resolution crystal structures of Sgt2_TPR alone and in complex with a C-terminal peptide PTVEEVD from heat shock protein, Ssa1. Using nuclear magnetic resonance spectroscopy and isothermal titration calorimetry, we demonstrate that Sgt2_TPR interacts with peptides corresponding to the C-termini of Ssa1, Hsc82, and Ybr137wp with similar binding modes and affinities.
Organizational Affiliation:
Department of Chemistry, King's College London, London, United Kingdom.