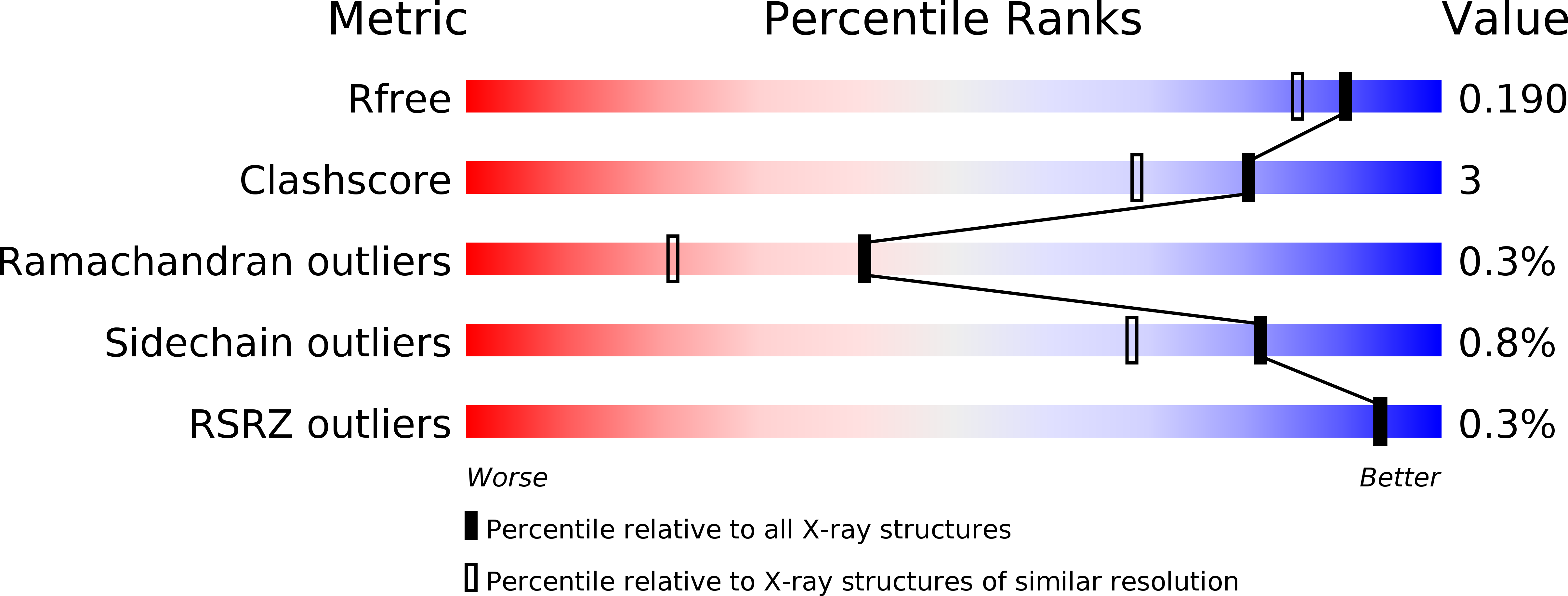
Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
Halland, N., Czech, J., Czechtizky, W., Evers, A., Follmann, M., Kohlmann, M., Schreuder, H.A., Kallus, C.(2016) J Med Chem 59: 9567-9573
- PubMed: 27749053
- DOI: https://doi.org/10.1021/acs.jmedchem.6b01276
- Primary Citation of Related Structures:
5LYD, 5LYF, 5LYI, 5LYL - PubMed Abstract:
Previously disclosed TAFIa inhibitors having a urea zinc-binding motif were used as the starting point for the development of a novel class of highly potent inhibitors having a sulfamide zinc-binding motif. High-resolution X-ray cocrystal structures were used to optimize the structures and reveal a highly unusual sulfamide configuration. A selected sulfamide was profiled in vitro and in vivo and displayed a promising ADMET profile.
Organizational Affiliation:
Sanofi R&D , Industriepark Höchst Building G838, D-65926 Frankfurt am Main, Germany.