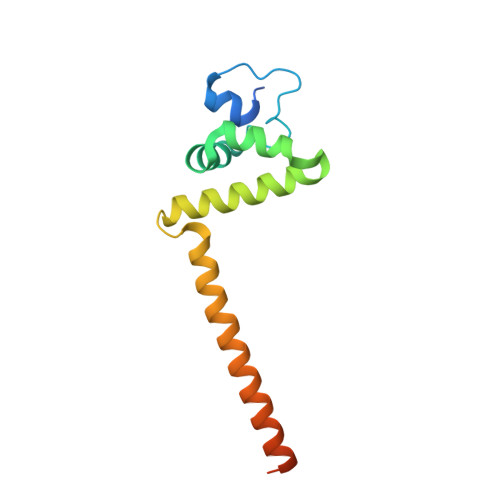
Structural analysis reveals a "molecular calipers" mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat.
Chen, W.F., Wei, X.B., Rety, S., Huang, L.Y., Liu, N.N., Dou, S.X., Xi, X.G.(2019) J Biol Chem 294: 142-156
- PubMed: 30425099
- DOI: https://doi.org/10.1074/jbc.RA118.003956
- Primary Citation of Related Structures:
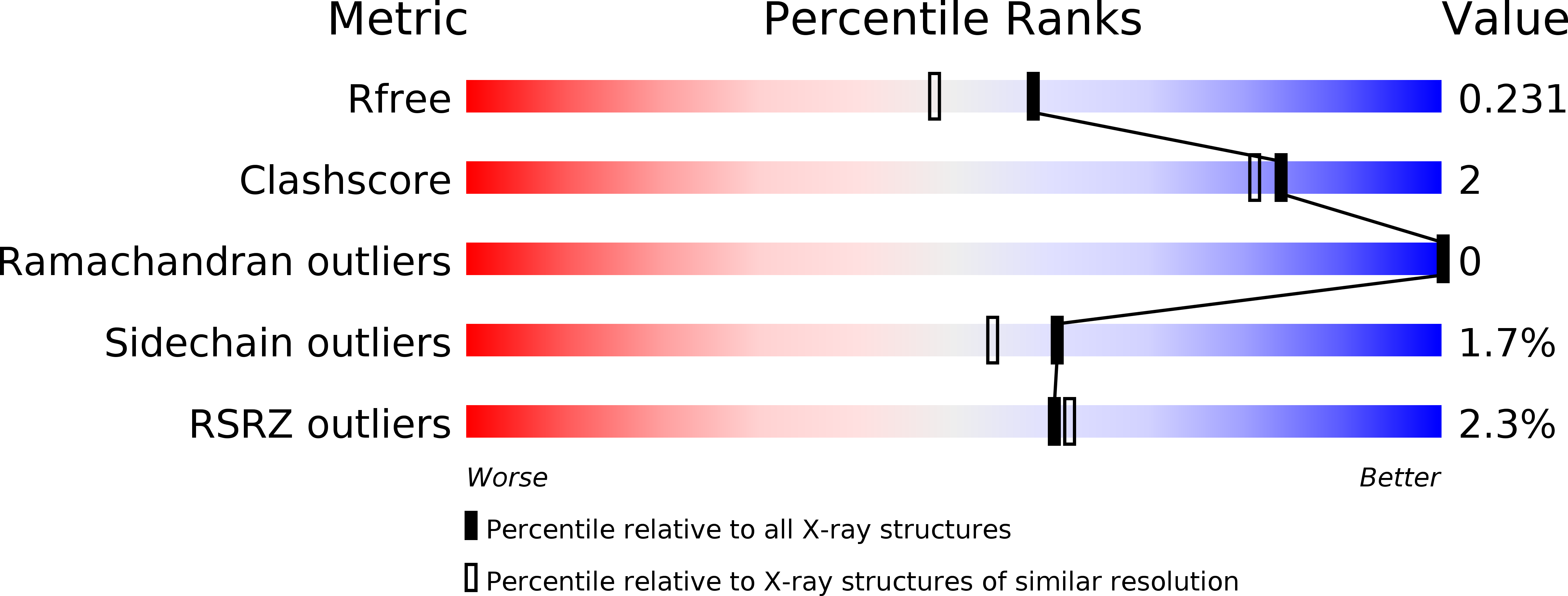
5LY0 - PubMed Abstract:
LATERAL ORGAN BOUNDARIES DOMAIN (LBD) proteins, a family of plant-specific transcription factors harboring a conserved Lateral Organ Boundaries (LOB) domain, are regulators of plant organ development. Recent studies have unraveled additional pivotal roles of the LBD protein family beyond defining lateral organ boundaries, such as pollen development and nitrogen metabolism. The structural basis for the molecular network of LBD-dependent processes remains to be deciphered. Here, we solved the first structure of the homodimeric LOB domain of Ramosa2 from wheat (TtRa2LD) to 1.9 Å resolution. Our crystal structure reveals structural features shared with other zinc-finger transcriptional factors, as well as some features unique to LBD proteins. Formation of the TtRa2LD homodimer relied on hydrophobic interactions of its coiled-coil motifs. Several specific motifs/domains of the LBD protein were also involved in maintaining its overall conformation. The intricate assembly within and between the monomers determined the precise spatial configuration of the two zinc fingers that recognize palindromic DNA sequences. Biochemical, molecular modeling, and small-angle X-ray scattering experiments indicated that dimerization is important for cooperative DNA binding and discrimination of palindromic DNA through a molecular calipers mechanism. Along with previously published data, this study enables us to establish an atomic-scale mechanistic model for LBD proteins as transcriptional regulators in plants.
Organizational Affiliation:
State Key Laboratory of Crop Stress Biology in Arid Areas, College of Life Sciences, Northwest A&F University, Yangling, Shaanxi 712100, China.