Structure and function of Aspergillus niger laccase McoG
Ferraroni, M., Westphal, A.H., Borsari, M., Tamayo-Ramos, J.A., Briganti, F., de Graaff, L.H., van Berkel, W.J.H.(2017) Biocatalysis
Experimental Data Snapshot
(2017) Biocatalysis
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Multicopper oxidase | 577 | Aspergillus niger | Mutation(s): 1 Gene Names: An08g08450 EC: 1.10.3.2 | 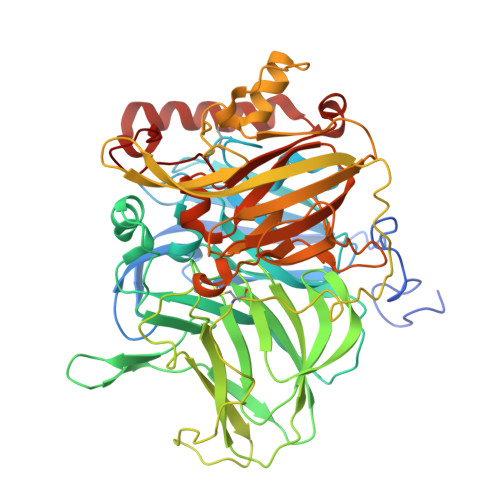 | |
UniProt | |||||
Find proteins for A2QS62 (Aspergillus niger (strain ATCC MYA-4892 / CBS 513.88 / FGSC A1513)) Explore A2QS62 Go to UniProtKB: A2QS62 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A2QS62 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | H [auth A], J [auth A], K [auth A], M [auth A], N [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| BMA Query on BMA | I [auth A], O [auth A] | beta-D-mannopyranose C6 H12 O6 WQZGKKKJIJFFOK-RWOPYEJCSA-N |  | ||
| MAN Query on MAN | L [auth A], P [auth A] | alpha-D-mannopyranose C6 H12 O6 WQZGKKKJIJFFOK-PQMKYFCFSA-N |  | ||
| CU Query on CU | D [auth A], E [auth A], F [auth A], G [auth A] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||
| PER Query on PER | Q [auth A] | PEROXIDE ION O2 ANAIPYUSIMHBEL-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 200.273 | α = 90 |
| b = 60.498 | β = 94.96 |
| c = 53.744 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XSCALE | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| REFMAC | phasing |