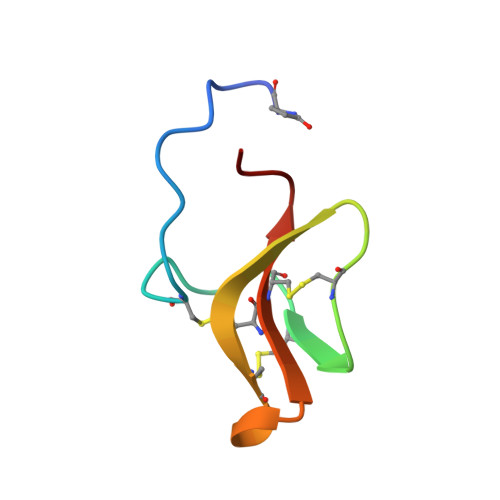
The Unusual Resistance of Avian Defensin AvBD7 to Proteolytic Enzymes Preserves Its Antibacterial Activity.
Bailleul, G., Kravtzoff, A., Joulin-Giet, A., Lecaille, F., Labas, V., Meudal, H., Loth, K., Teixeira-Gomes, A.P., Gilbert, F.B., Coquet, L., Jouenne, T., Bromme, D., Schouler, C., Landon, C., Lalmanach, G., Lalmanach, A.C.(2016) PLoS One 11: e0161573-e0161573
- PubMed: 27561012
- DOI: https://doi.org/10.1371/journal.pone.0161573
- Primary Citation of Related Structures:
5LCS - PubMed Abstract:
Defensins are frontline peptides of mucosal immunity in the animal kingdom, including birds. Their resistance to proteolysis and their ensuing ability to maintain antimicrobial potential remains questionable and was therefore investigated. We have shown by bottom-up mass spectrometry analysis of protein extracts that both avian beta-defensins AvBD2 and AvBD7 were ubiquitously distributed along the chicken gut. Cathepsin B was found by immunoblotting in jejunum, ileum, caecum, and caecal tonsils, while cathepsins K, L, and S were merely identified in caecal tonsils. Hydrolysis product of AvBD2 and AvBD7 incubated with a panel of proteases was analysed by RP-HPLC, mass spectrometry and antimicrobial assays. AvBD2 and AvBD7 were resistant to serine proteases and to cathepsins D and H. Conversely cysteine cathepsins B, K, L, and S degraded AvBD2 and abolished its antibacterial activity. Only cathepsin K cleaved AvBD7 and released Ile4-AvBD7, a N-terminal truncated natural peptidoform of AvBD7 that displayed antibacterial activity. Besides the 3-stranded antiparallel beta-sheet typical of beta-defensins, structural analysis of AvBD7 by two-dimensional NMR spectroscopy highlighted the restricted accessibility of the C-terminus embedded by the N-terminal region and gave a formal evidence of a salt bridge (Asp9-Arg12) that could account for proteolysis resistance. The differential susceptibility of avian defensins to proteolysis opens intriguing questions about a distinctive role in the mucosal immunity against pathogen invasion.
Organizational Affiliation:
ISP, INRA, Université François Rabelais de Tours, UMR 1282, Nouzilly, France.