Crystal Structure of a Clostripain (BT_0727) from Bacteroides thetaiotaomicron ATCC 29148 in Complex with Peptide Inhibitor BTN-VLTK-AOMK
Wolan, D.W., Roncase, E.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Clostripain-related protein | 143 | Bacteroides thetaiotaomicron VPI-5482 | Mutation(s): 0 Gene Names: BT_0727 | 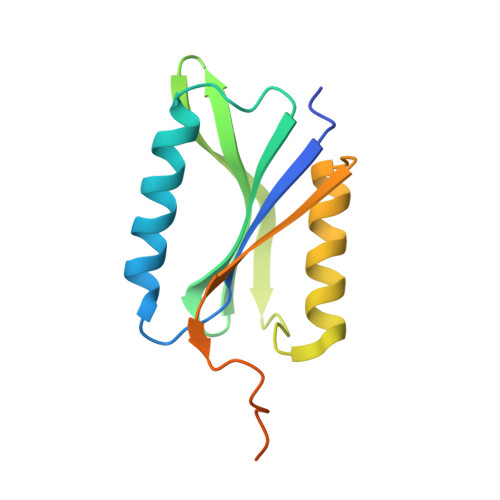 | |
UniProt | |||||
Find proteins for Q8A9T8 (Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50)) Explore Q8A9T8 Go to UniProtKB: Q8A9T8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8A9T8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Clostripain-related protein | 229 | Bacteroides thetaiotaomicron VPI-5482 | Mutation(s): 0 Gene Names: BT_0727 | 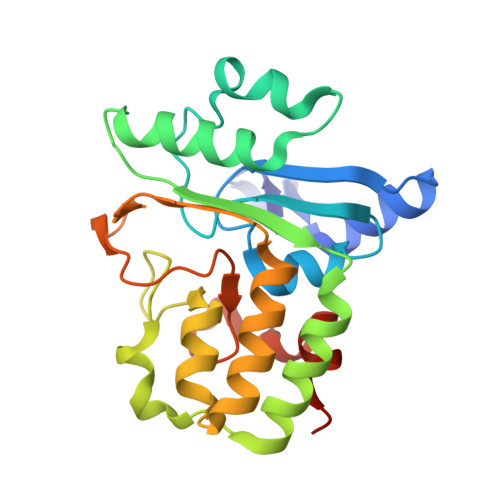 | |
UniProt | |||||
Find proteins for Q8A9T8 (Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50)) Explore Q8A9T8 Go to UniProtKB: Q8A9T8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8A9T8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptide Inhibitor BTN-VLTK-AOMK | 5 | synthetic construct | Mutation(s): 0 | 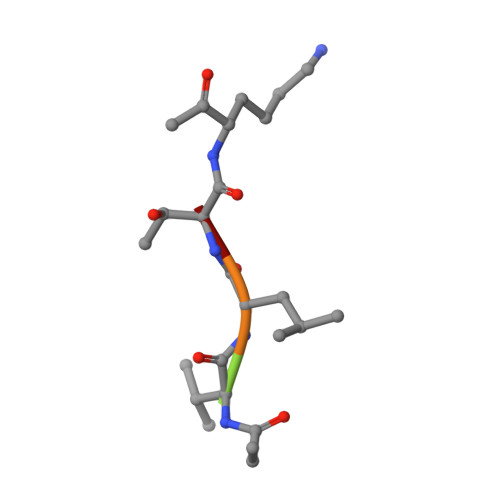 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_002239 Query on PRD_002239 | C | Peptide Inhibitor BTN-VLTK-AOMK | Peptide-like / Inhibitor |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 79.207 | α = 90 |
| b = 102.885 | β = 90 |
| c = 84.186 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 3U54GM094586-03S1 |