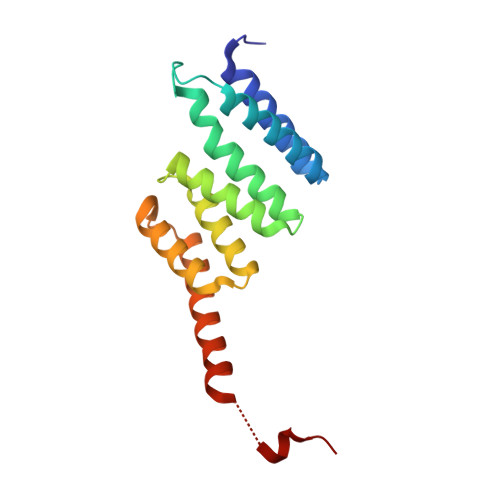
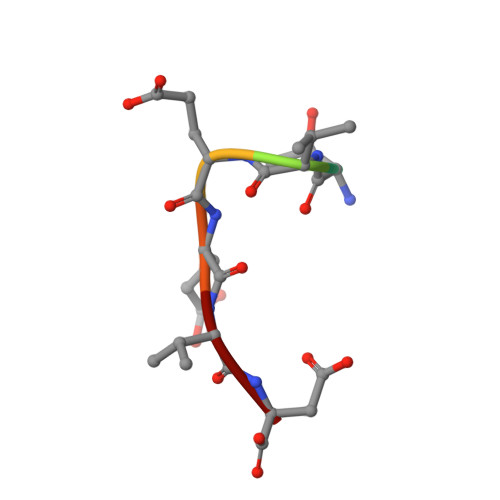
Two alternative binding mechanisms connect the protein translocation Sec71-Sec72 complex with heat shock proteins.
Tripathi, A., Mandon, E.C., Gilmore, R., Rapoport, T.A.(2017) J Biol Chem 292: 8007-8018
- PubMed: 28286332
- DOI: https://doi.org/10.1074/jbc.M116.761122
- Primary Citation of Related Structures:
5L0W, 5L0Y - PubMed Abstract:
The biosynthesis of many eukaryotic proteins requires accurate targeting to and translocation across the endoplasmic reticulum membrane. Post-translational protein translocation in yeast requires both the Sec61 translocation channel, and a complex of four additional proteins: Sec63, Sec62, Sec71, and Sec72. The structure and function of these proteins are largely unknown. This pathway also requires the cytosolic Hsp70 protein Ssa1, but whether Ssa1 associates with the translocation machinery to target protein substrates to the membrane is unclear. Here, we use a combined structural and biochemical approach to explore the role of Sec71-Sec72 subcomplex in post-translational protein translocation. To this end, we report a crystal structure of the Sec71-Sec72 complex, which revealed that Sec72 contains a tetratricopeptide repeat (TPR) domain that is anchored to the endoplasmic reticulum membrane by Sec71. We also determined the crystal structure of this TPR domain with a C-terminal peptide derived from Ssa1, which suggests how Sec72 interacts with full-length Ssa1. Surprisingly, Ssb1, a cytoplasmic Hsp70 that binds ribosome-associated nascent polypeptide chains, also binds to the TPR domain of Sec72, even though it lacks the TPR-binding C-terminal residues of Ssa1. We demonstrate that Ssb1 binds through its ATPase domain to the TPR domain, an interaction that leads to inhibition of nucleotide exchange. Taken together, our results suggest that translocation substrates can be recruited to the Sec71-Sec72 complex either post-translationally through Ssa1 or co-translationally through Ssb1.
Organizational Affiliation:
From the Howard Hughes Medical Institute and the Department of Cell Biology, Harvard Medical School, Boston, Massachusetts 02115 and arati_tripathi@hms.harvard.edu.