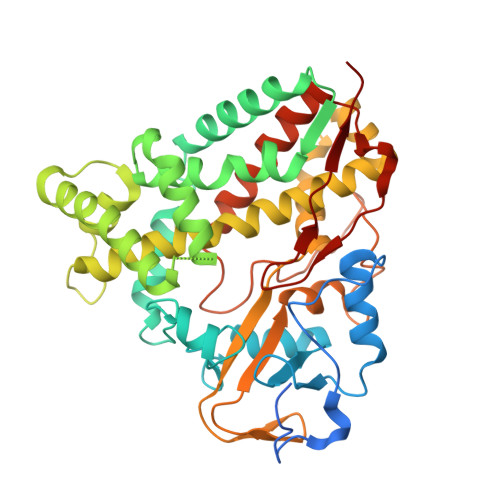
X-ray crystal structure of cytochrome P450 monooxygenase CYP101J2 from Sphingobium yanoikuyae strain B2.
Unterweger, B., Drinkwater, N., Johanesen, P., Lyras, D., Dumsday, G.J., McGowan, S.(2017) Proteins 85: 945-950
- PubMed: 27936485
- DOI: https://doi.org/10.1002/prot.25227
- Primary Citation of Related Structures:
5KYO - PubMed Abstract:
The cytochrome P450 monooxygenases (P450s) catalyze a vast array of oxygenation reactions that can be useful in biocatalytic applications. CYP101J2 from Sphingobium yanoikuyae is a P450 that catalyzes the hydroxylation of 1,8-cineole. Here we report the crystallization and X-ray structure elucidation of recombinant CYP101J2 to 1.8 Å resolution. The CYP101J2 structure shows the canonical P450-fold and has an open conformation in the absence of substrate. Analysis of the structure revealed that CYP101J2, in the absence of substrate, forms a well-ordered substrate-binding channel that suggests a unique form of substrate guidance in comparison to other bacterial 1,8-cineole-hydroxylating P450 enzymes. Proteins 2017; 85:945-950. © 2016 Wiley Periodicals, Inc.
Organizational Affiliation:
Infection and Immunity Program, Biomedicine Discovery Institute, Department of Microbiology, Monash University, Clayton, Victoria, 3800.