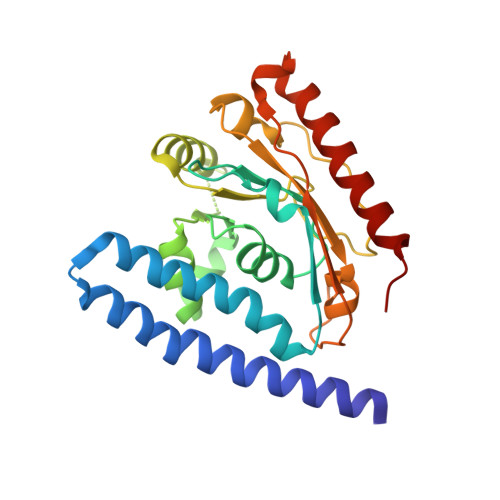
A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer.
Robertson, R.M., Yao, J., Gajewski, S., Kumar, G., Martin, E.W., Rock, C.O., White, S.W.(2017) Nat Struct Mol Biol 24: 666-671
- PubMed: 28714993
- DOI: https://doi.org/10.1038/nsmb.3436
- Primary Citation of Related Structures:
5KYM - PubMed Abstract:
Phosphatidic acid (PA), the central intermediate in membrane phospholipid synthesis, is generated by two acyltransferases in a pathway conserved in all life forms. The second step in this pathway is catalyzed by 1-acyl-sn-glycerol-3-phosphate acyltransferase, called PlsC in bacteria. Here we present the crystal structure of PlsC from Thermotoga maritima, revealing an unusual hydrophobic/aromatic N-terminal two-helix motif linked to an acyltransferase αβ-domain that contains the catalytic HX 4 D motif. PlsC dictates the acyl chain composition of the 2-position of phospholipids, and the acyl chain selectivity 'ruler' is an appropriately placed and closed hydrophobic tunnel. We confirmed this by site-directed mutagenesis and membrane composition analysis of Escherichia coli cells that expressed mutant PlsC. Molecular dynamics (MD) simulations showed that the two-helix motif represents a novel substructure that firmly anchors the protein to one leaflet of the membrane. This binding mode allows the PlsC active site to acylate lysophospholipids within the membrane bilayer by using soluble acyl donors.
Organizational Affiliation:
Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, Tennessee, USA.