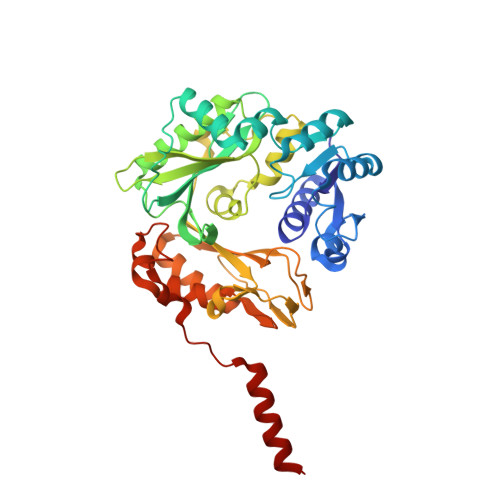
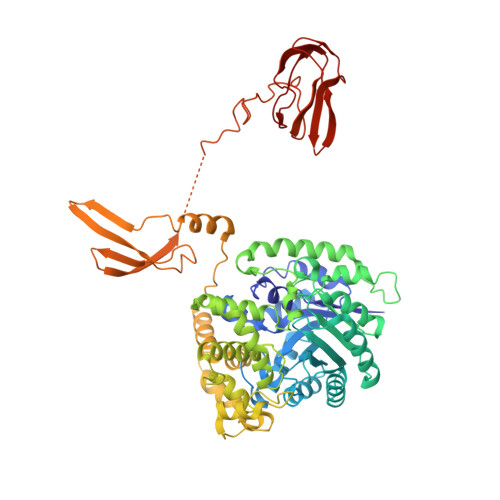
A distinct holoenzyme organization for two-subunit pyruvate carboxylase.
Choi, P.H., Jo, J., Lin, Y.C., Lin, M.H., Chou, C.Y., Dietrich, L.E., Tong, L.(2016) Nat Commun 7: 12713-12713
- PubMed: 27708276
- DOI: https://doi.org/10.1038/ncomms12713
- Primary Citation of Related Structures:
5KS8 - PubMed Abstract:
Pyruvate carboxylase (PC) has important roles in metabolism and is crucial for virulence for some pathogenic bacteria. PC contains biotin carboxylase (BC), carboxyltransferase (CT) and biotin carboxyl carrier protein (BCCP) components. It is a single-chain enzyme in eukaryotes and most bacteria, and functions as a 500 kD homo-tetramer. In contrast, PC is a two-subunit enzyme in a collection of Gram-negative bacteria, with the α subunit containing the BC and the β subunit the CT and BCCP domains, and it is believed that the holoenzyme has α 4 β 4 stoichiometry. We report here the crystal structures of a two-subunit PC from Methylobacillus flagellatus. Surprisingly, our structures reveal an α 2 β 4 stoichiometry, and the overall architecture of the holoenzyme is strikingly different from that of the homo-tetrameric PCs. Biochemical and mutagenesis studies confirm the stoichiometry and other structural observations. Our functional studies in Pseudomonas aeruginosa show that its two-subunit PC is important for colony morphogenesis.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York 10027, USA.