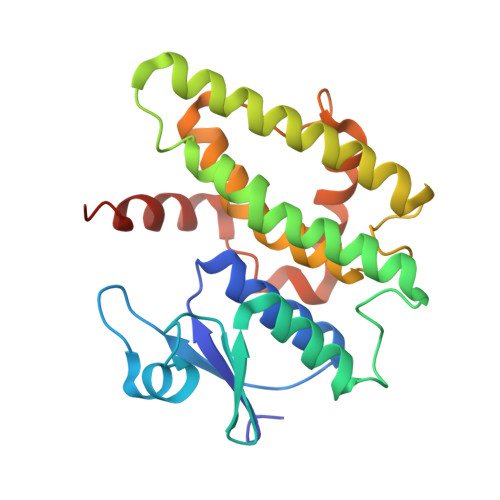
Insights into ligand binding to a glutathione S-transferase from mango: Structure, thermodynamics and kinetics.
Valenzuela-Chavira, I., Contreras-Vergara, C.A., Arvizu-Flores, A.A., Serrano-Posada, H., Lopez-Zavala, A.A., Garcia-Orozco, K.D., Hernandez-Paredes, J., Rudino-Pinera, E., Stojanoff, V., Sotelo-Mundo, R.R., Islas-Osuna, M.A.(2017) Biochimie 135: 35-45
- PubMed: 28104507
- DOI: https://doi.org/10.1016/j.biochi.2017.01.005
- Primary Citation of Related Structures:
5G5E, 5G5F, 5KEJ - PubMed Abstract:
We studied a mango glutathione S-transferase (GST) (Mangifera indica) bound to glutathione (GSH) and S-hexyl glutathione (GSX). This GST Tau class (MiGSTU) had a molecular mass of 25.5 kDa. MiGSTU Michaelis-Menten kinetic constants were determined for their substrates obtaining a K m , V max and k cat for CDNB of 0.792 mM, 80.58 mM min -1 and 68.49 s -1 respectively and 0.693 mM, 105.32 mM min -1 and 89.57 s -1 , for reduced GSH respectively. MiGSTU had a micromolar affinity towards GSH (5.2 μM) or GSX (7.8 μM). The crystal structure of the MiGSTU in apo or bound to GSH or GSX generated a model that explains the thermodynamic signatures of binding and showed the importance of enthalpic-entropic compensation in ligand binding to Tau-class GST enzymes.
Organizational Affiliation:
Laboratorio de Estructura Biomolecular, Centro de Investigación en Alimentación y Desarrollo, A.C. (CIAD), Hermosillo, Sonora 83304, Mexico; Laboratorio de Genética Molecular de Plantas, Centro de Investigación en Alimentación y Desarrollo, A.C. (CIAD), Hermosillo, Sonora 83304, Mexico.