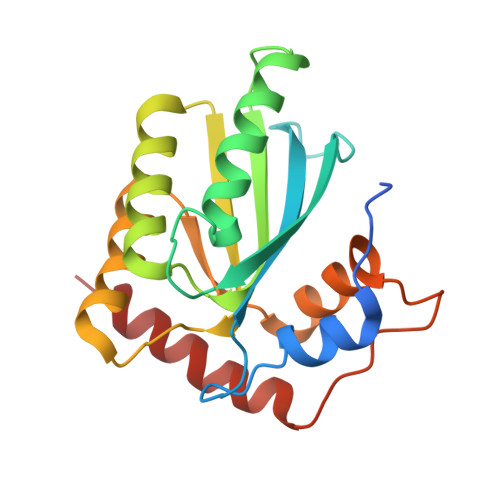
Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Xiao, X., Li, S.X., Yang, H., Chen, X.S.(2016) Nat Commun 7: 12193-12193
- PubMed: 27480941
- DOI: https://doi.org/10.1038/ncomms12193
- Primary Citation of Related Structures:
5K81, 5K82, 5K83 - PubMed Abstract:
APOBEC3G (A3G) is a potent restriction factor of HIV-1. The N-terminal domain of A3G (A3G-CD1) is responsible for oligomerization and nucleic acid binding, both of which are essential for anti-HIV activity. As a countermeasure, HIV-1 viral infectivity factor (Vif) binds A3G-CD1 to mediate A3G degradation. The structural basis for the functions of A3G-CD1 remains elusive. Here, we report the crystal structures of a primate A3G-CD1 (rA3G-CD1) alone and in complex with single-stranded DNA (ssDNA). rA3G-CD1 shares a conserved core structure with the previously determined catalytic APOBECs, but displays unique features for surface charge, dimerization and nucleic acid binding. Its co-crystal structure with ssDNA reveals how the conformations of loops and residues surrounding the Zn-coordinated centre (Zn-centre) change upon DNA binding. The dimerization interface of rA3G-CD1 is important for oligomerization, nucleic acid binding and Vif-mediated degradation. These findings elucidate the molecular basis of antiviral mechanism and HIV-Vif targeting of A3G.
Organizational Affiliation:
Genetic, Molecular and Cellular Biology Program, Keck School of Medicine, University of Southern California, Los Angeles, California 90089, USA.