Structural analysis of the activation-induced deoxycytidine deaminase required in immunoglobulin diversification.
Pham, P., Afif, S.A., Shimoda, M., Maeda, K., Sakaguchi, N., Pedersen, L.C., Goodman, M.F.(2016) DNA Repair (Amst) 43: 48-56
- PubMed: 27258794
- DOI: https://doi.org/10.1016/j.dnarep.2016.05.029
- Primary Citation of Related Structures:
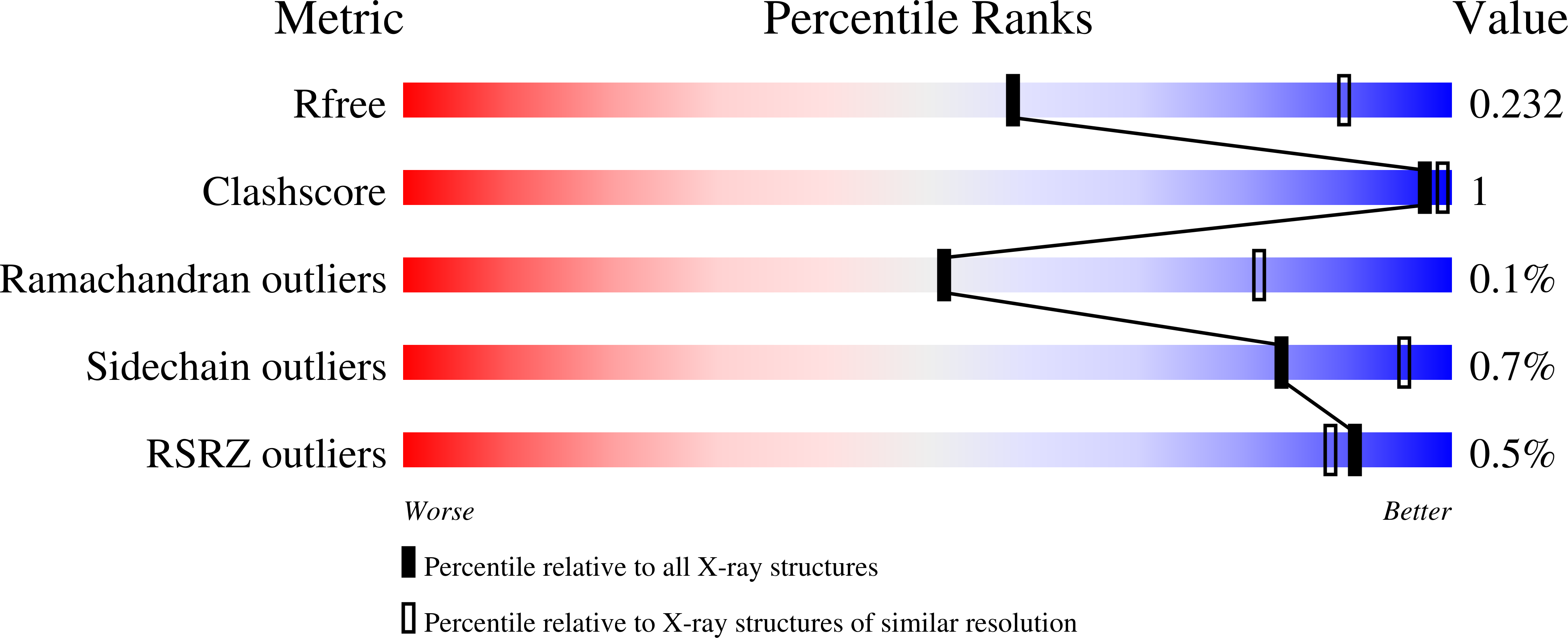
5JJ4 - PubMed Abstract:
Activation-induced deoxycytidine deaminase (AID) initiates somatic hypermutation (SHM) and class-switch recombination (CSR) by deaminating C→U during transcription of Ig-variable (V) and Ig-switch (S) region DNA, which is essential to produce high-affinity antibodies. Here we report the crystal structure of a soluble human AID variant at 2.8Å resolution that favors targeting WRC motifs (W=A/T, R=A/G) in vitro, and executes Ig V SHM in Ramos B-cells. A specificity loop extending away from the active site to accommodate two purine bases next to C, differs significantly in sequence, length, and conformation from APOBEC proteins Apo3A and Apo3G, which strongly favor pyrimidines at -1 and -2 positions. Individual amino acid contributions to specificity and processivity were measured in relation to a proposed ssDNA binding cleft. This study provides a structural basis for residue contributions to DNA scanning properties unique to AID, and for disease mutations in human HIGM-2 syndrome.
Organizational Affiliation:
Department of Biological Sciences, University of Southern California, Los Angeles, CA, 90089, United States.